Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
Arif, S.M., Varshney, U., Vijayan, M.(2017) J Struct Biol 199: 165-176
- PubMed: 28705712
- DOI: https://doi.org/10.1016/j.jsb.2017.07.002
- Primary Citation of Related Structures:
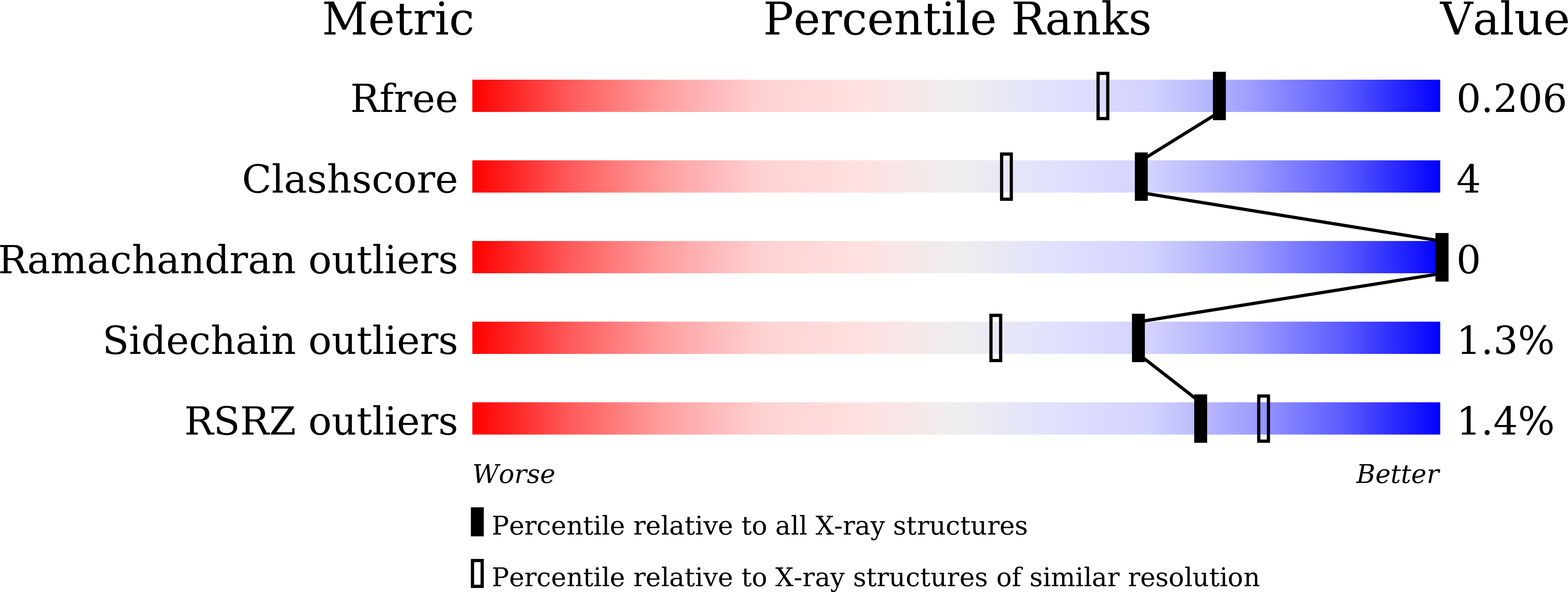
5XD1, 5XD2, 5XD3, 5XD4, 5XD5 - PubMed Abstract:
Diadenosine polyphosphates (Ap n A, n=2-6), particularly Ap 4 A, are involved in several important physiological processes. The substantial sequence identity of the Nudix hydrolase domain (domain 1) of Mycobacterium smegmatis MutT1 (MsMutT1) with a known Ap 4 A hydrolase suggested that MsMutT1 could also hydrolyse diadenosine polyphosphates. Biochemical experiments yielded results in conformity with this suggestion, with Ap 4 A as the best among the substrates. ATP is a product in all experiments; small amounts of ADP were also observed in the experiments involving Ap 4 A and Ap 6 A. Hydrolysis was inhibited by fluoride ions in all cases. The mechanism of action and its inhibition in relation to Ap n A were explored through the X-ray analysis of the crystals of the MsMutT1 complexes with Ap 5 A; Ap 5 A and MnCl 2 ; Ap 4 A; ATP; and ATP.NaF.MgCl 2 . The aggregation pattern of molecules in the first four crystals is similar to that found in a majority of MsMutT1-NTP crystals. Substrate molecules occupy the primary binding site and ATP occupies a site at an intermolecular interface, in the first two. ATP occupies both the sites in the third and fourth crystal. The protein-ligand interactions observed in these crystal structures lead to an explanation of the molecular mechanism of hydrolysis of Ap n A by MsMutT1. The fifth crystal exhibits a new packing arrangement. The structure of the complex provides an explanation for the fluoride inhibition of the activity of the enzyme. It would thus appear that MutT1 has a major role involving the hydrolysis of diadenosine polyphosphates, which could be elucidated at the molecular level.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560 012, India.