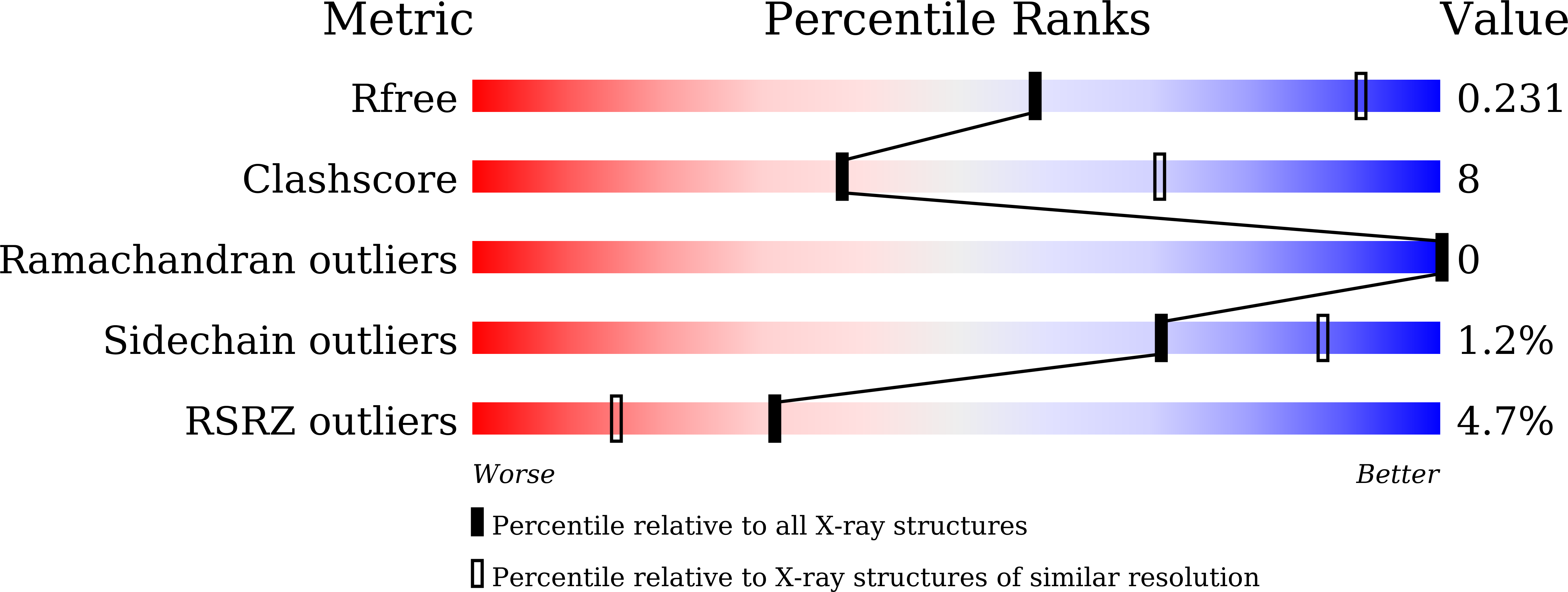
Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Chai, N., Li, W.X., Wang, J., Wang, Z.X., Yang, S.M., Wu, J.W.(2015) Nucleic Acids Res 43: 9051-9064
- PubMed: 26304548
- DOI: https://doi.org/10.1093/nar/gkv848
- Primary Citation of Related Structures:
5X6G, 5X6H, 5X6M - PubMed Abstract:
Smad proteins are important intracellular mediators of TGF-β signalling, which transmit signals directly from cell surface receptors to the nucleus. The MH1 domain of Smad plays a key role in DNA recognition. Two types of DNA sequence were identified as Smad binding motifs: the Smad binding element (SBE) and the GC-rich sequence. Here we report the first crystal structure of the Smad5 MH1 domain in complex with the GC-rich sequence. Compared with the Smad5-MH1/SBE complex structure, the Smad5 MH1 domain contacts the GC-rich site with the same β-hairpin, but the detailed interaction modes are different. Conserved β-hairpin residues make base specific contacts with the minimal GC-rich site, 5'-GGC-3'. The assembly of Smad5-MH1 on the GC-rich DNA also results in distinct DNA conformational changes. Moreover, the crystal structure of Smad5-MH1 in complex with a composite DNA sequence demonstrates that the MH1 domain is targeted to each binding site (GC-rich or SBE) with modular binding modes, and the length of the DNA spacer affects the MH1 assembly. In conclusion, our work provides the structural basis for the recognition and binding specificity of the Smad MH1 domain with the DNA targets.
Organizational Affiliation:
MOE Key Laboratory of Protein Science, School of Life Sciences, Tsinghua University, Beijing 100084, China.