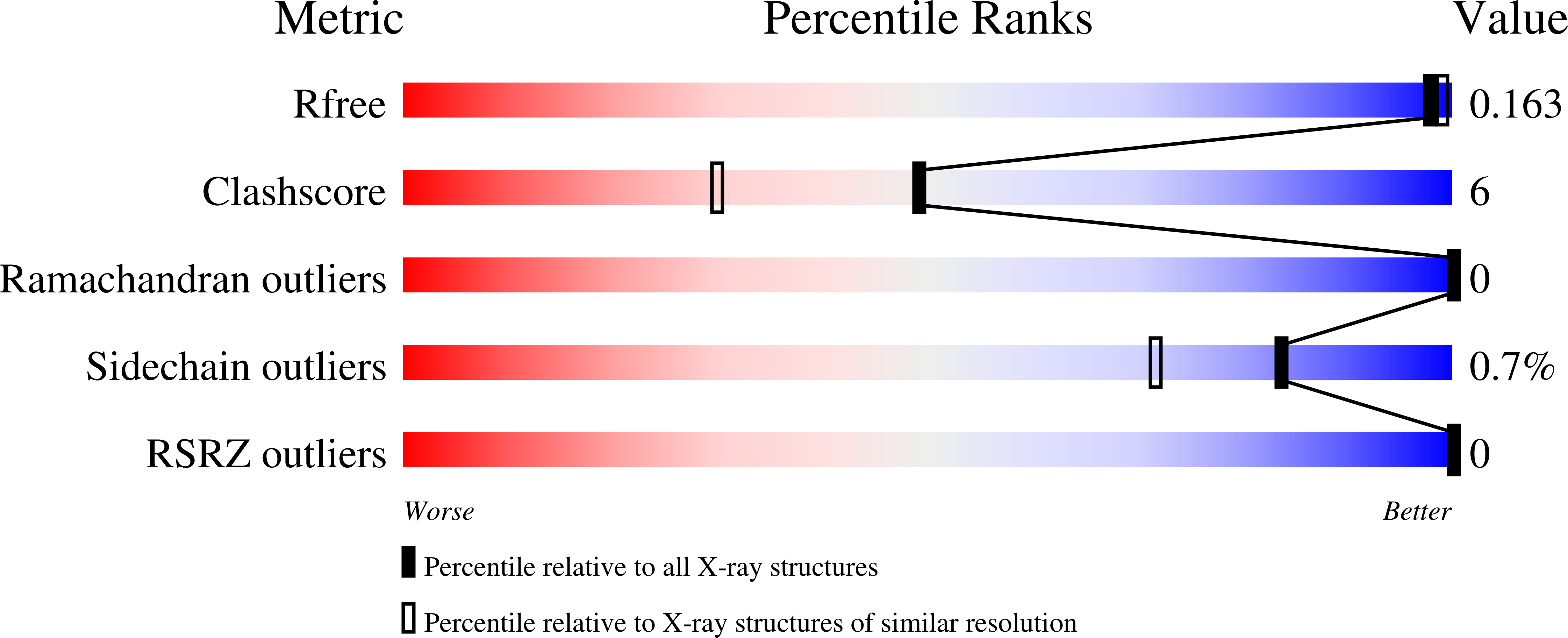
First biochemical and crystallographic characterization of a fast-performing ferritin from a marine invertebrate.
De Meulenaere, E., Bailey, J.B., Tezcan, F.A., Deheyn, D.D.(2017) Biochem J 474: 4193-4206
- PubMed: 29127253
- DOI: https://doi.org/10.1042/BCJ20170681
- Primary Citation of Related Structures:
5WPN - PubMed Abstract:
Ferritin, a multimeric cage-like enzyme, is integral to iron metabolism across all phyla through the sequestration and storage of iron through efficient ferroxidase activity. While ferritin sequences from ∼900 species have been identified, crystal structures from only 50 species have been reported, the majority from bacterial origin. We recently isolated a secreted ferritin from the marine invertebrate Chaetopterus sp. (parchment tube worm), which resides in muddy coastal seafloors. Here, we present the first ferritin from a marine invertebrate to be crystallized and its biochemical characterization. The initial ferroxidase reaction rate of recombinant Chaetopterus ferritin (ChF) is 8-fold faster than that of recombinant human heavy-chain ferritin (HuHF). To our knowledge, this protein exhibits the fastest catalytic performance ever described for a ferritin variant. In addition to the high-velocity ferroxidase activity, ChF is unique in that it is secreted by Chaetopterus in a bioluminescent mucus. Previous work has linked the availability of Fe 2+ to this long-lived bioluminescence, suggesting a potential function for the secreted ferritin. Comparative biochemical analyses indicated that both ChF and HuHF showed similar behavior toward changes in pH, temperature, and salt concentration. Comparison of their crystal structures shows no significant differences in the catalytic sites. Notable differences were found in the residues that line both 3-fold and 4-fold pores, potentially leading to increased flexibility, reduced steric hindrance, or a more efficient pathway for Fe 2+ transportation to the ferroxidase site. These suggested residues could contribute to the understanding of iron translocation through the ferritin shell to the ferroxidase site.
Organizational Affiliation:
Marine Biology Research Division, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA 92037, U.S.A.