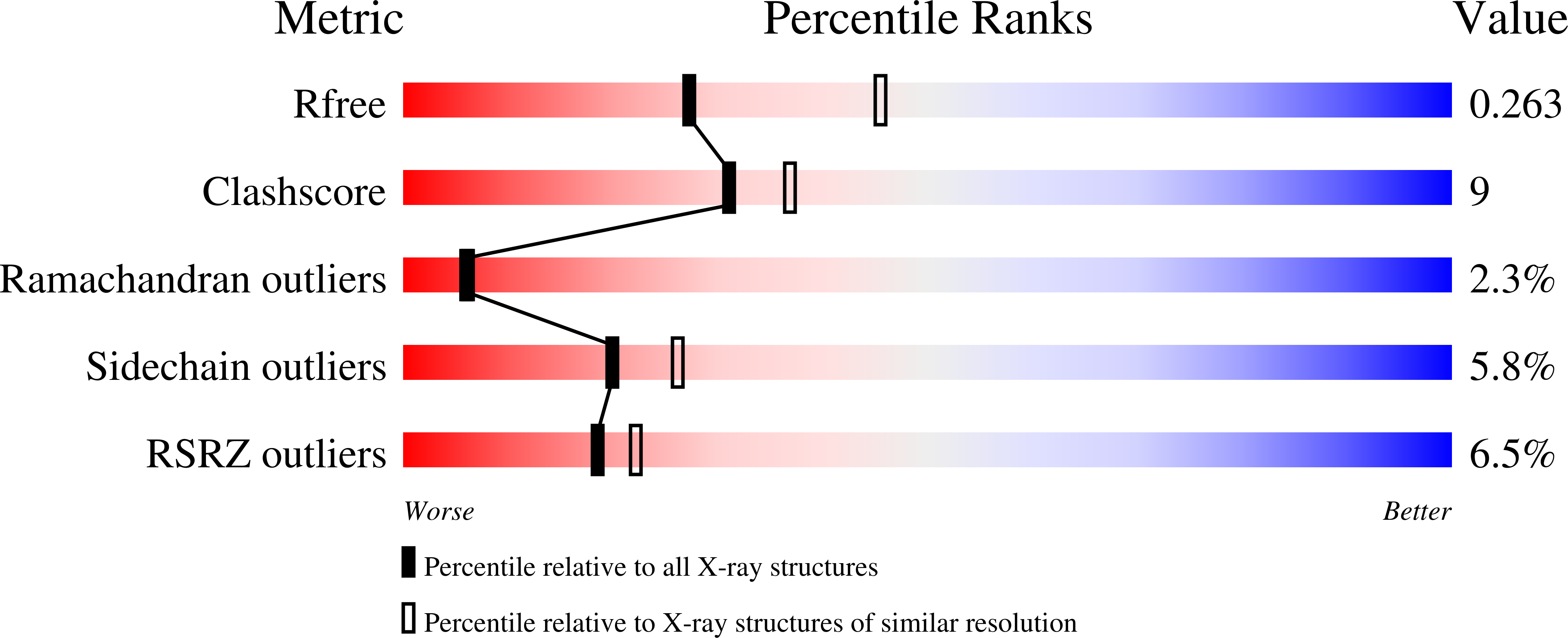
Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Jacquet, P., Hiblot, J., Daude, D., Bergonzi, C., Gotthard, G., Armstrong, N., Chabriere, E., Elias, M.(2017) Sci Rep 7: 16745-16745
- PubMed: 29196634
- DOI: https://doi.org/10.1038/s41598-017-16841-0
- Primary Citation of Related Structures:
5VRI, 5VRK, 5VSA, 5W3U, 5W3W, 5W3Z - PubMed Abstract:
The redesign of enzyme active sites to alter their function or specificity is a difficult yet appealing challenge. Here we used a structure-based design approach to engineer the lactonase SsoPox from Sulfolobus solfataricus into a phosphotriesterase. The five best variants were characterized and their structure was solved. The most active variant, αsD6 (V27A-Y97W-L228M-W263M) demonstrates a large increase in catalytic efficiencies over the wild-type enzyme, with increases of 2,210-fold, 163-fold, 58-fold, 16-fold against methyl-parathion, malathion, ethyl-paraoxon, and methyl-paraoxon, respectively. Interestingly, the best mutants are also capable of degrading fensulfothion, which is reported to be an inhibitor for the wild-type enzyme, as well as others that are not substrates of the starting template or previously reported W263 mutants. The broad specificity of these engineered variants makes them promising candidates for the bioremediation of organophosphorus compounds. Analysis of their structures reveals that the increase in activity mainly occurs through the destabilization of the active site loop involved in substrate binding, and it has been observed that the level of disorder correlates with the width of the enzyme specificity spectrum. This finding supports the idea that active site conformational flexibility is essential to the acquisition of broader substrate specificity.
Organizational Affiliation:
CNRS UMR 7278, IRD198, INSERM U1095, APHM, Institut Hospitalier Universitaire Méditerranée-Infection, Aix-Marseille Université, 19-21 Bd Jean Moulin, 13005, Marseille, France.