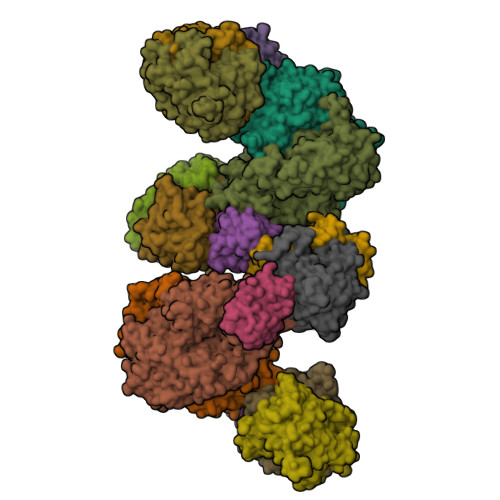
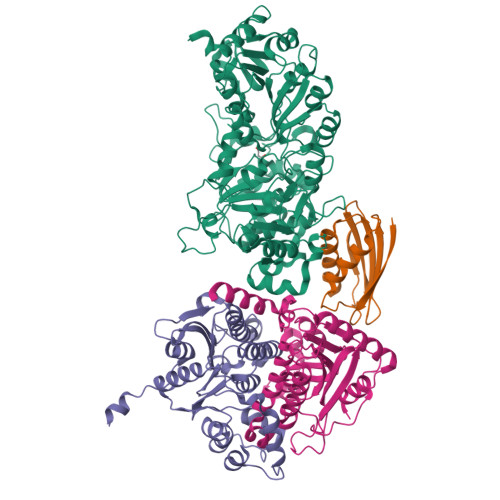
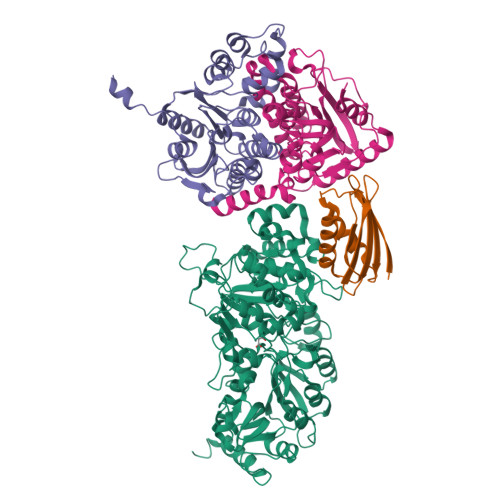
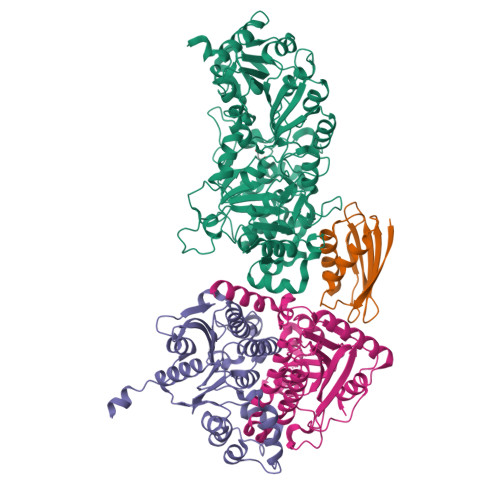
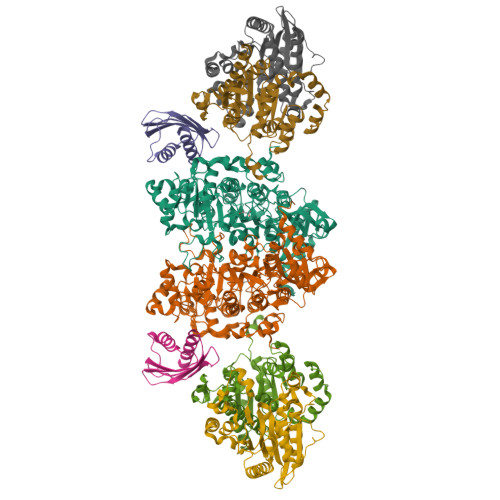
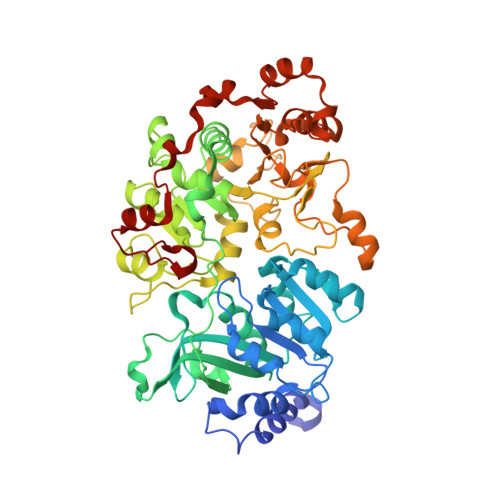
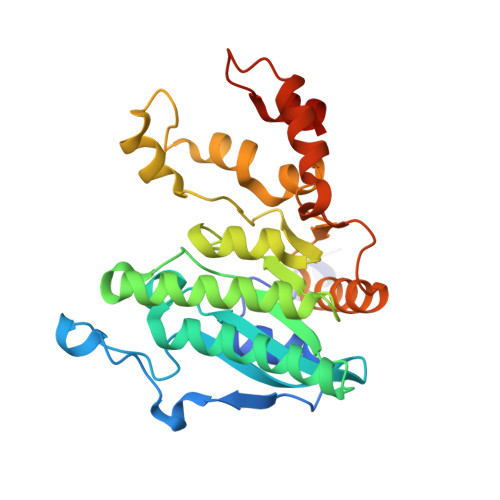
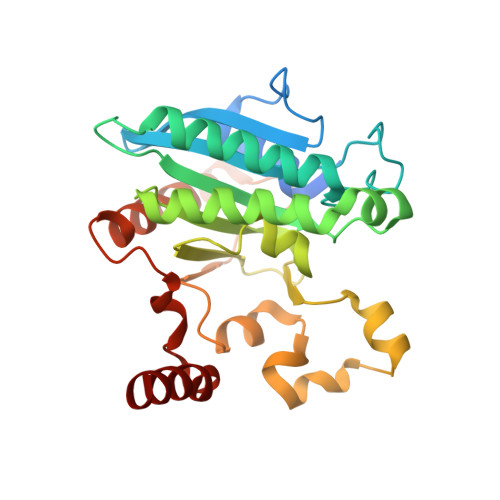
Crystal structure of a Pseudomonas malonate decarboxylase holoenzyme hetero-tetramer.
Maderbocus, R., Fields, B.L., Hamilton, K., Luo, S., Tran, T.H., Dietrich, L.E.P., Tong, L.(2017) Nat Commun 8: 160-160
- PubMed: 28757619
- DOI: https://doi.org/10.1038/s41467-017-00233-z
- Primary Citation of Related Structures:
5VIP, 5VIT, 5VJ1 - PubMed Abstract:
Pseudomonas species and other aerobic bacteria have a biotin-independent malonate decarboxylase that is crucial for their utilization of malonate as the sole carbon and energy source. The malonate decarboxylase holoenzyme contains four subunits, having an acyl-carrier protein (MdcC subunit) with a distinct prosthetic group, as well as decarboxylase (MdcD-MdcE) and acyl-carrier protein transferase (MdcA) catalytic activities. Here we report the crystal structure of a Pseudomonas malonate decarboxylase hetero-tetramer, as well as biochemical and functional studies based on the structural information. We observe a malonate molecule in the active site of MdcA and we also determine the structure of malonate decarboxylase with CoA in the active site of MdcD-MdcE. Both structures provide molecular insights into malonate decarboxylase catalysis. Mutations in the hetero-tetramer interface can abolish holoenzyme formation. Mutations in the hetero-tetramer interface and the active sites can abolish Pseudomonas aeruginosa growth in a defined medium with malonate as the sole carbon source.Some aerobic bacteria contain a biotin-independent malonate decarboxylase (MDC), which allows them to use malonate as the sole carbon source. Here, the authors present the crystal structure of a Pseudomonas MDC and give insights into its catalytic mechanism and function.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, NY, 10027, USA.