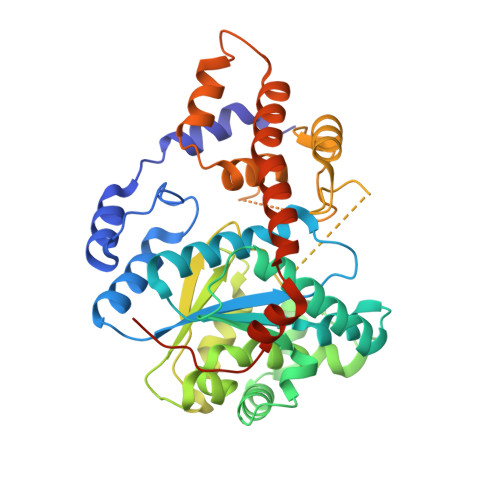
An alternative conformation of human TrpRS suggests a role of zinc in activating non-enzymatic function.
Xu, X., Zhou, H., Zhou, Q., Hong, F., Vo, M.N., Niu, W., Wang, Z., Xiong, X., Nakamura, K., Wakasugi, K., Schimmel, P., Yang, X.L.(2018) RNA Biol 15: 649-658
- PubMed: 28910573
- DOI: https://doi.org/10.1080/15476286.2017.1377868
- Primary Citation of Related Structures:
5UJI, 5UJJ - PubMed Abstract:
Tryptophanyl-tRNA synthetase (TrpRS) in vertebrates contains a N-terminal extension in front of the catalytic core. Proteolytic removal of the N-terminal 93 amino acids gives rise to T2-TrpRS, which has potent anti-angiogenic activity mediated through its extracellular interaction with VE-cadherin. Zinc has been shown to have anti-angiogenic effects and can bind to human TrpRS. However, the connection between zinc and the anti-angiogenic function of TrpRS has not been explored. Here we report that zinc binding can induce structural relaxation in human TrpRS to facilitate the proteolytic generation of a T2-TrpRS-like fragment. The zinc-binding site is likely to be contained within T2-TrpRS, and the zinc-bound conformation of T2-TrpRS is mimicked by mutation H130R. We determined the crystal structure of H130R T2-TrpRS at 2.8 Å resolution, which reveals drastically different conformation from that of wild-type (WT) T2-TrpRS. The conformational change creates larger binding surfaces for VE-cadherin as suggested by molecular dynamic simulations. Surface plasmon resonance analysis indicates more than 50-fold increase in binding affinity of H130R T2-TrpRS for VE-cadherin, compared to WT T2-TrpRS. The enhanced interaction is also confirmed by a cell-based binding analysis. These results suggest that zinc plays an important role in activating TrpRS for angiogenesis regulation.
Organizational Affiliation:
a Department of Molecular Medicine , The Scripps Research Institute , La Jolla , CA , USA.