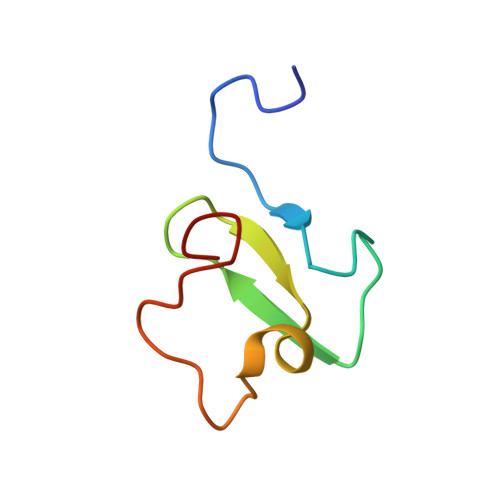
PHF20 Readers Link Methylation of Histone H3K4 and p53 with H4K16 Acetylation.
Klein, B.J., Wang, X., Cui, G., Yuan, C., Botuyan, M.V., Lin, K., Lu, Y., Wang, X., Zhao, Y., Bruns, C.J., Mer, G., Shi, X., Kutateladze, T.G.(2016) Cell Rep 17: 1158-1170
- PubMed: 27760318
- DOI: https://doi.org/10.1016/j.celrep.2016.09.056
- Primary Citation of Related Structures:
5TAB, 5TBN - PubMed Abstract:
PHF20 is a core component of the lysine acetyltransferase complex MOF (male absent on the first)-NSL (non-specific lethal) that generates the major epigenetic mark H4K16ac and is necessary for transcriptional regulation and DNA repair. The role of PHF20 in the complex remains elusive. Here, we report on functional coupling between methylation readers in PHF20. We show that the plant homeodomain (PHD) finger of PHF20 recognizes dimethylated lysine 4 of histone H3 (H3K4me2) and represents an example of a native reader that selects for this modification. Biochemical and structural analyses help to explain this selectivity and the preference of Tudor2, another reader in PHF20, for dimethylated p53. Binding of the PHD finger to H3K4me2 is required for histone acetylation, accumulation of PHF20 at target genes, and transcriptional activation. Together, our findings establish a unique PHF20-mediated link between MOF histone acetyltransferase (HAT), p53, and H3K4me2, and suggest a model for rapid spreading of H4K16ac-enriched open chromatin.
Organizational Affiliation:
Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045, USA.