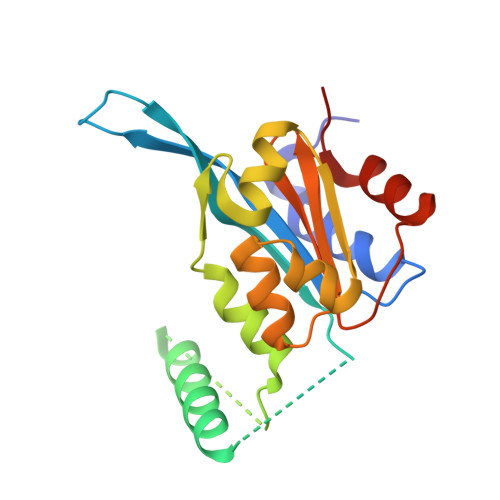
Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Zhang, F., Ke, J., Zhang, L., Chen, R., Sugimoto, K., Howe, G.A., Xu, H.E., Zhou, M., He, S.Y., Melcher, K.(2017) Proc Natl Acad Sci U S A 114: 1720-1725
- PubMed: 28137867
- DOI: https://doi.org/10.1073/pnas.1616938114
- Primary Citation of Related Structures:
5T0F, 5T0Q - PubMed Abstract:
Jasmonate ZIM-domain (JAZ) transcriptional repressors play a key role in regulating jasmonate (JA) signaling in plants. Below a threshold concentration of jasmonoyl isoleucine (JA-Ile), the active form of JA, the C-terminal Jas motif of JAZ proteins binds MYC transcription factors to repress JA signaling. With increasing JA-Ile concentration, the Jas motif binds to JA-Ile and the COI1 subunit of the SCF COI1 E3 ligase, which mediates ubiquitination and proteasomal degradation of JAZ repressors, resulting in derepression of MYC transcription factors. JA signaling subsequently becomes desensitized, in part by feedback induction of JAZ splice variants that lack the C-terminal Jas motif but include an N-terminal cryptic MYC-interaction domain (CMID). The CMID sequence is dissimilar to the Jas motif and is incapable of recruiting SCF COI1 , allowing CMID-containing JAZ splice variants to accumulate in the presence of JA and to re-repress MYC transcription factors as an integral part of reestablishing signal homeostasis. The mechanism by which the CMID represses MYC transcription factors remains elusive. Here we describe the crystal structure of the MYC3-CMID JAZ10 complex. In contrast to the Jas motif, which forms a single continuous helix when bound to MYC3, the CMID adopts a loop-helix-loop-helix architecture with modular interactions with both the Jas-binding groove and the backside of the Jas-interaction domain of MYC3. This clamp-like interaction allows the CMID to bind MYC3 tightly and block access of MED25 (a subunit of the Mediator coactivator complex) to the MYC3 transcriptional activation domain, shedding light on the enigmatic mechanism by which JAZ splice variants desensitize JA signaling.
Organizational Affiliation:
College of Plant Protection, Nanjing Agricultural University, 210095, Nanjing, Jiangsu Province, China.