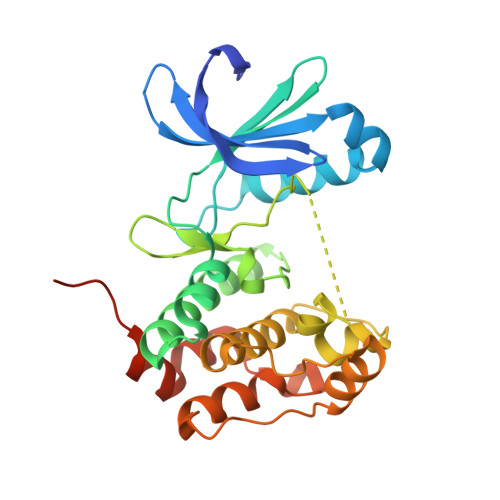
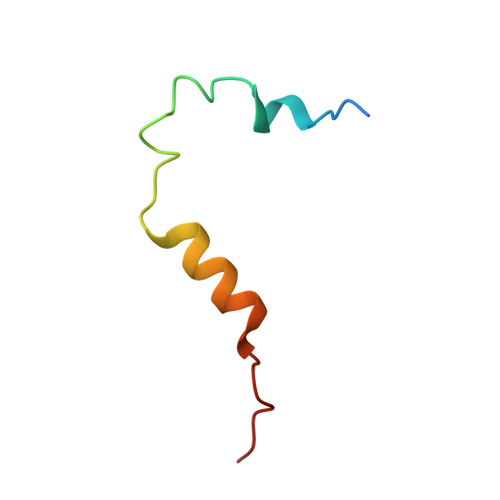
Mitotic spindle association of TACC3 requires Aurora-A-dependent stabilization of a cryptic alpha-helix.
Burgess, S.G., Mukherjee, M., Sabir, S., Joseph, N., Gutierrez-Caballero, C., Richards, M.W., Huguenin-Dezot, N., Chin, J.W., Kennedy, E.J., Pfuhl, M., Royle, S.J., Gergely, F., Bayliss, R.(2018) EMBO J 37
- PubMed: 29510984
- DOI: https://doi.org/10.15252/embj.201797902
- Primary Citation of Related Structures:
5ODT - PubMed Abstract:
Aurora-A regulates the recruitment of TACC3 to the mitotic spindle through a phospho-dependent interaction with clathrin heavy chain (CHC). Here, we describe the structural basis of these interactions, mediated by three motifs in a disordered region of TACC3. A hydrophobic docking motif binds to a previously uncharacterized pocket on Aurora-A that is blocked in most kinases. Abrogation of the docking motif causes a delay in late mitosis, consistent with the cellular distribution of Aurora-A complexes. Phosphorylation of Ser558 engages a conformational switch in a second motif from a disordered state, needed to bind the kinase active site, into a helical conformation. The helix extends into a third, adjacent motif that is recognized by a helical-repeat region of CHC, not a recognized phospho-reader domain. This potentially widespread mechanism of phospho-recognition provides greater flexibility to tune the molecular details of the interaction than canonical recognition motifs that are dominated by phosphate binding.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology, School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, UK.