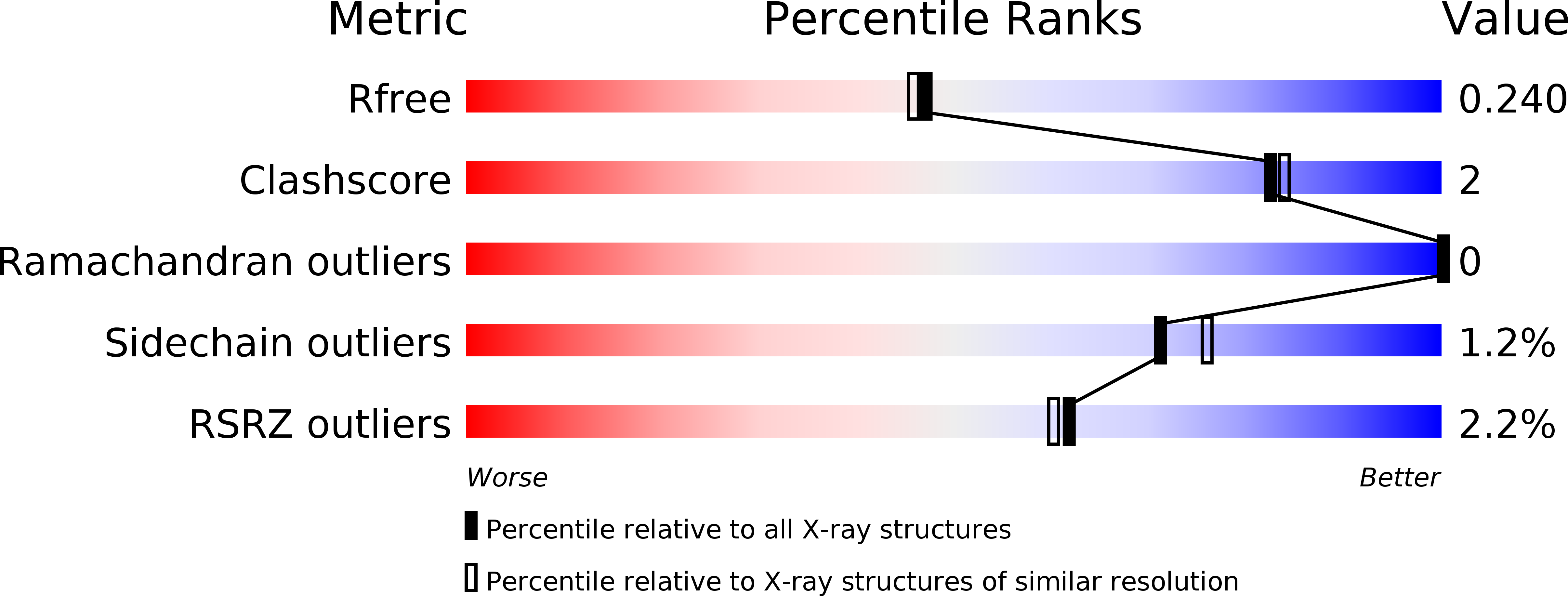
Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Narwal, M., Jemth, A.S., Gustafsson, R., Almlof, I., Warpman Berglund, U., Helleday, T., Stenmark, P.(2018) Biochemistry 57: 593-603
- PubMed: 29281266
- DOI: https://doi.org/10.1021/acs.biochem.7b01163
- Primary Citation of Related Structures:
5MZE, 5MZF, 5MZG, 6EHH - PubMed Abstract:
MTH1 hydrolyzes oxidized nucleoside triphosphates, thereby sanitizing the nucleotide pool from oxidative damage. This prevents incorporation of damaged nucleotides into DNA, which otherwise would lead to mutations and cell death. The high level of reactive oxygen species in cancer cells leads to a higher level of oxidized nucleotides in cancer cells compared to that in nonmalignant cells, making cancer cells more dependent on MTH1 for survival. The possibility of specifically targeting cancer cells by inhibiting MTH1 has highlighted MTH1 as a promising cancer target. The progression of MTH1 inhibitors into the clinic requires animal studies, and knowledge of species differences in the potency of inhibitors is vitally important. We here show that the human MTH1 inhibitor TH588 is approximately 20-fold less potent with respect to inhibition of mouse MTH1 than the human, rat, pig, and dog MTH1 proteins are. We present the crystal structures of mouse MTH1 in complex with TH588 and dog MTH1 and elucidate the structural and sequence basis for the observed difference in affinity for TH588. We identify amino acid residue 116 in MTH1 as an important determinant of TH588 affinity. Furthermore, we present the structure of mouse MTH1 in complex with the substrate 8-oxo-dGTP. The crystal structures provide insight into the high degree of structural conservation between MTH1 proteins from different organisms and provide a detailed view of interactions between MTH1 and the inhibitor, revealing that minute structural differences can have a large impact on affinity and specificity.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University , S-106 91 Stockholm, Sweden.