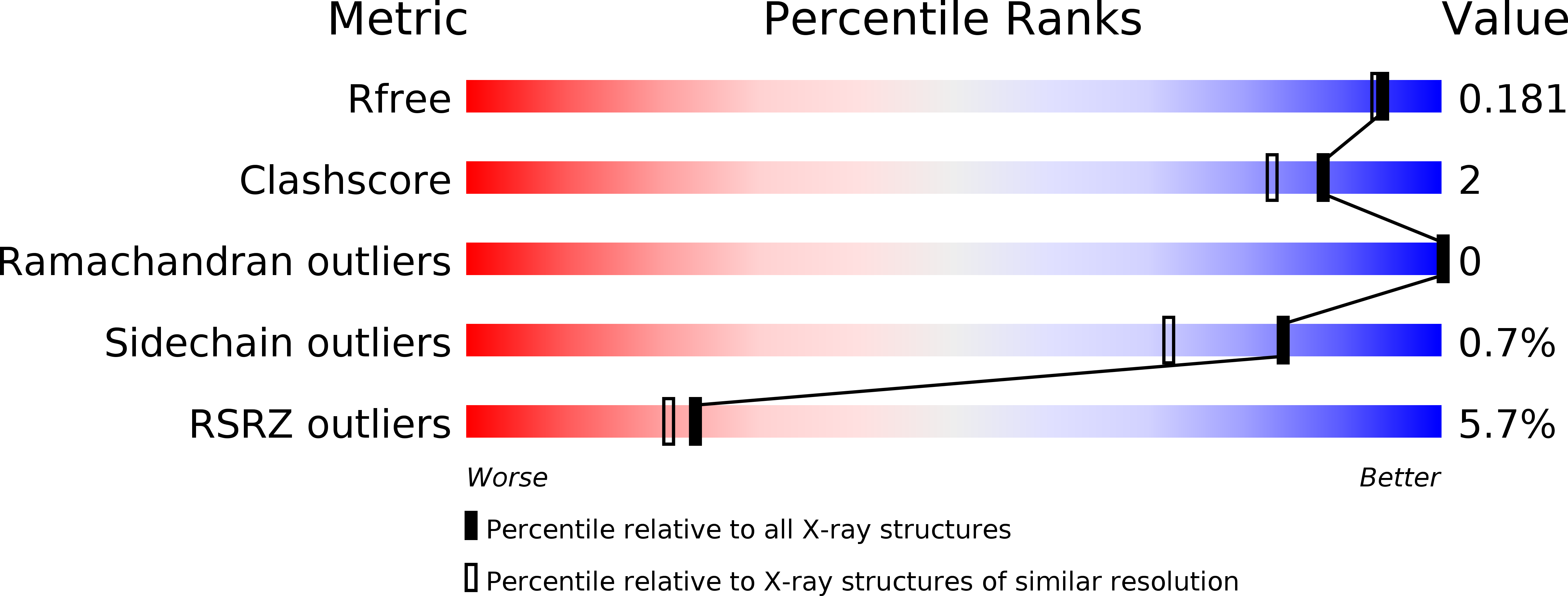
Target Identification of Mycobacterium tuberculosis Phenotypic Hits Using a Concerted Chemogenomic, Biophysical, and Structural Approach.
Mugumbate, G., Mendes, V., Blaszczyk, M., Sabbah, M., Papadatos, G., Lelievre, J., Ballell, L., Barros, D., Abell, C., Blundell, T.L., Overington, J.P.(2017) Front Pharmacol 8: 681-681
- PubMed: 29018348
- DOI: https://doi.org/10.3389/fphar.2017.00681
- Primary Citation of Related Structures:
5MXV, 5MYL, 5MYM, 5MYN, 5MYR, 5MYS, 5MYT, 5MYW - PubMed Abstract:
Mycobacterium phenotypic hits are a good reservoir for new chemotypes for the treatment of tuberculosis. However, the absence of defined molecular targets and modes of action could lead to failure in drug development. Therefore, a combination of ligand-based and structure-based chemogenomic approaches followed by biophysical and biochemical validation have been used to identify targets for Mycobacterium tuberculosis phenotypic hits. Our approach identified EthR and InhA as targets for several hits, with some showing dual activity against these proteins. From the 35 predicted EthR inhibitors, eight exhibited an IC 50 below 50 μM against M. tuberculosis EthR and three were confirmed to be also simultaneously active against InhA. Further hit validation was performed using X-ray crystallography yielding eight new crystal structures of EthR inhibitors. Although the EthR inhibitors attain their activity against M. tuberculosis by hitting yet undefined targets, these results provide new lead compounds that could be further developed to be used to potentiate the effect of EthA activated pro-drugs, such as ethionamide, thus enhancing their bactericidal effect.
Organizational Affiliation:
European Molecular Biology Laboratory, European Bioinformatics Institute, Cambridge, United Kingdom.