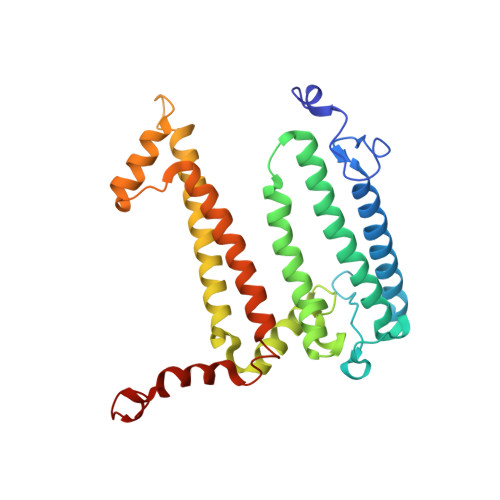
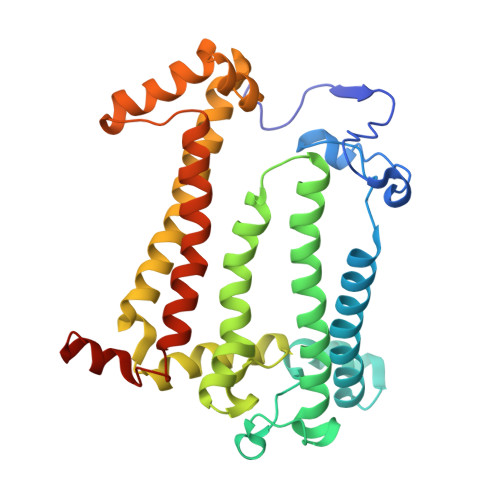
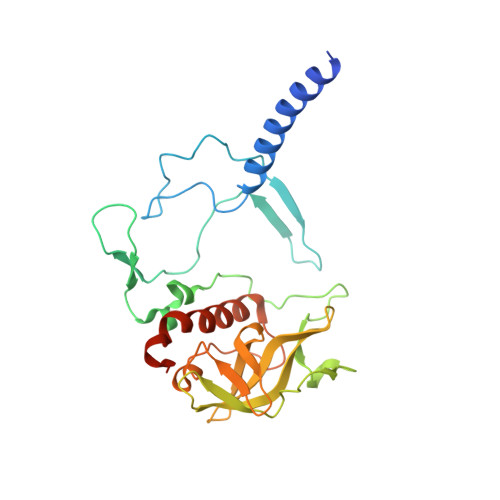
On the mechanism of ubiquinone mediated photocurrent generation by a reaction center based photocathode.
Friebe, V.M., Swainsbury, D.J., Fyfe, P.K., van der Heijden, W., Jones, M.R., Frese, R.N.(2016) Biochim Biophys Acta 1857: 1925-1934
- PubMed: 27687473
- DOI: https://doi.org/10.1016/j.bbabio.2016.09.011
- Primary Citation of Related Structures:
5LRI, 5LSE - PubMed Abstract:
Upon photoexcitation, the reaction center (RC) pigment-proteins that facilitate natural photosynthesis achieve a metastable separation of electrical charge among the embedded cofactors. Because of the high quantum efficiency of this process, there is a growing interest in their incorporation into biohybrid materials for solar energy conversion, bioelectronics and biosensing. Multiple bioelectrochemical studies have shown that reaction centers from various photosynthetic organisms can be interfaced with diverse electrode materials for the generation of photocurrents, but many mechanistic aspects of native protein functionality in a non-native environment is unknown. In vivo, RC's catalyse ubiquinone-10 reduction, protonation and exchange with other lipid phase ubiquinone-10s via protein-controlled spatial orientation and protein rearrangement. In contrast, the mechanism of ubiquinone-0 reduction, used to facilitate fast RC turnover in an aqueous photoelectrochemical cell (PEC), may not proceed via the same pathway as the native cofactor. In this report we show truncation of the native isoprene tail results in larger RC turnover rates in a PEC despite the removal of the tail's purported role of ubiquinone headgroup orientation and binding. Through the use of reaction centers with single or double mutations, we also show the extent to which two-electron/two-proton ubiquinone chemistry that operates in vivo also underpins the ubiquinone-0 reduction by surface-adsorbed RCs in a PEC. This reveals that only the ubiquinone headgroup is critical to the fast turnover of the RC in a PEC and provides insight into design principles for the development of new biophotovoltaic cells and biosensors.
Organizational Affiliation:
Department of Physics and Astronomy, LaserLaB Amsterdam, VU University Amsterdam, De Boelelaan 1081, Amsterdam 1081, HV, The Netherlands.