Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Ebert, C., Simon, N., Schneider, S., Carell, T.(2017) Chembiochem 18: 1379-1382
- PubMed: 28444956
- DOI: https://doi.org/10.1002/cbic.201700169
- Primary Citation of Related Structures:
5LCL, 5LCM - PubMed Abstract:
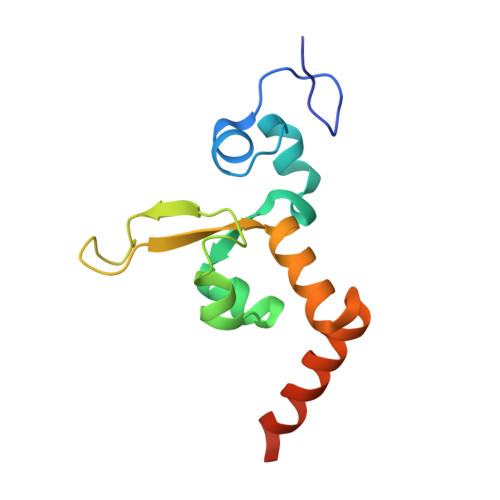
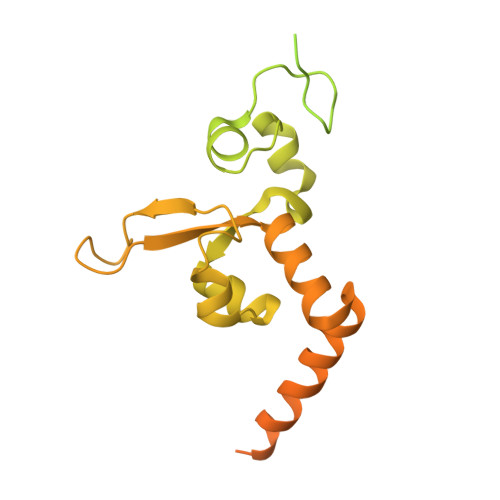
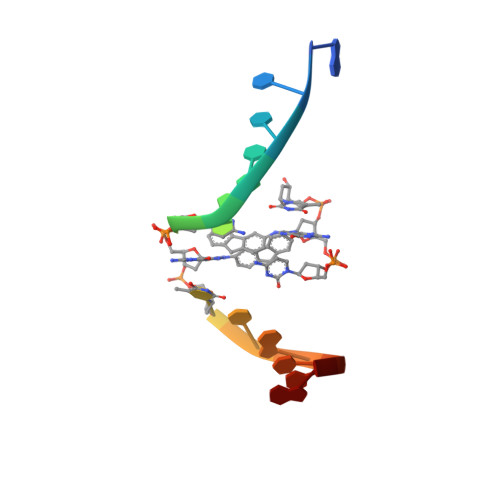
Aromatic amines are strongly carcinogenic. They are activated in the liver to give reactive nitrenium ions that react with nucleobases within the DNA duplex. The reaction occurs predominantly at the C8 position of the dG base, thereby giving C8-acetyl-aryl- or C8-aryl-dG adducts in an electrophilic aromatic substitution reaction. Alternatively, reaction with the exocyclic 2-NH 2 group is observed. Although the C8 adducts retain base-pairing properties, base pairing is strongly compromised in the case of the N 2 adducts. Here we show crystal structures of two DNA lesions, N 2 -acetylnaphthyl-dG and C8-fluorenyl-dG, within a DNA duplex recognized by the repair protein Rad14. The structures confirm that two molecules of the repair protein recognize the lesion and induce a 72 or 78° kink at the site of the damage. Importantly, the same overall kinked structure is induced by binding of the repair proteins, although the structurally different lesions result in distinct stacking interactions of the lesions within the duplex. The results suggest that the repair protein XPA/Rad14 is a sensor that recognizes flexibility. The protein converts the information that structurally different lesions are present in the duplex into a unifying sharply kinked recognition motif.
Organizational Affiliation:
Center for Integrated Protein Science at the Department of Chemistry, Ludwig-Maximilians-Universität München, Butenandtstrasse 5-13, 81377, München, Germany.