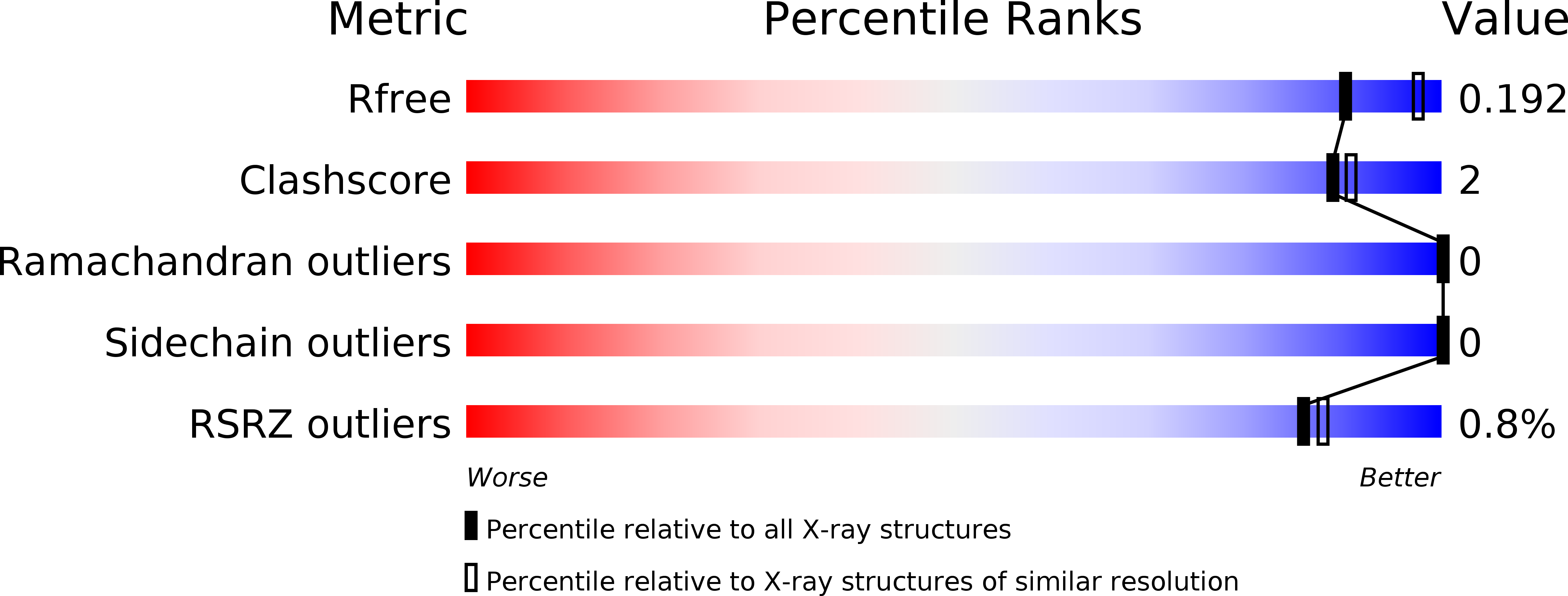
The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex.
Sammak, S., Allen, M.D., Hamdani, N., Bycroft, M., Zinzalla, G.(2018) FEBS J 285: 4165-4180
- PubMed: 30222246
- DOI: https://doi.org/10.1111/febs.14660
- Primary Citation of Related Structures:
5L7A, 5L7B - PubMed Abstract:
c-MYC and the SWI/SNF chromatin remodeling complex act as master regulators of transcription, and play a key role in human cancer. Although they are known to interact, the molecular details of their interaction are lacking. We have determined the structure of the RPT1 region of the INI1/hSNF5/BAF47/SMARCB1 subunit of the SWI/SNF complex that acts as a c-MYC-binding domain, and have localized the interaction regions on both INI1 and on the c-MYC:MAX heterodimer. c-MYC interacts with a highly conserved groove on INI1, while INI1 binds to the c-MYC helix-loop-helix region. The binding site overlaps with the c-MYC DNA-binding region, and we show that binding of INI1 and E-box DNA to c-MYC:MAX are mutually exclusive.
Organizational Affiliation:
Microbiology, Tumor and Cell Biology (MTC) Department, Karolinska Institutet, Stockholm, Sweden.