Direct visualization of a Fe(IV)-OH intermediate in a heme enzyme.
Kwon, H., Basran, J., Casadei, C.M., Fielding, A.J., Schrader, T.E., Ostermann, A., Devos, J.M., Aller, P., Blakeley, M.P., Moody, P.C., Raven, E.L.(2016) Nat Commun 7: 13445-13445
- PubMed: 27897163
- DOI: https://doi.org/10.1038/ncomms13445
- Primary Citation of Related Structures:
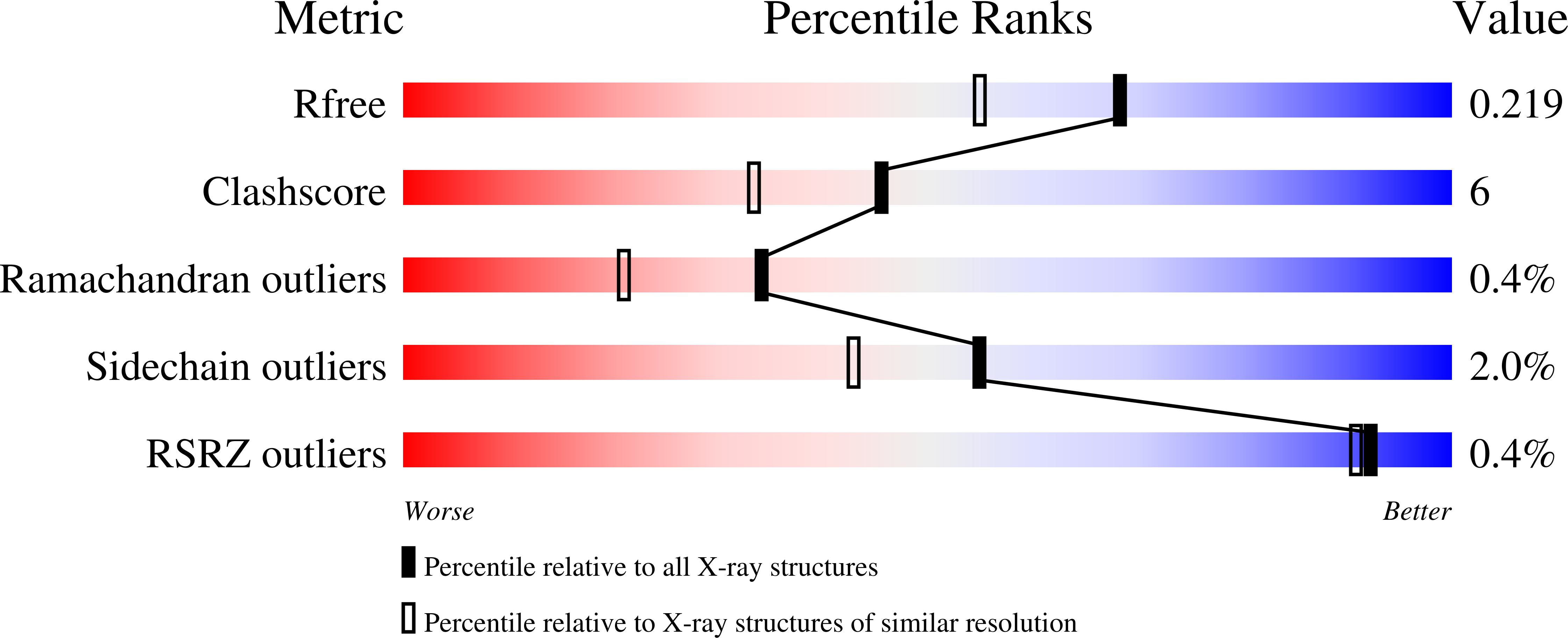
5JPR, 5JQR - PubMed Abstract:
Catalytic heme enzymes carry out a wide range of oxidations in biology. They have in common a mechanism that requires formation of highly oxidized ferryl intermediates. It is these ferryl intermediates that provide the catalytic engine to drive the biological activity. Unravelling the nature of the ferryl species is of fundamental and widespread importance. The essential question is whether the ferryl is best described as a Fe(IV)=O or a Fe(IV)-OH species, but previous spectroscopic and X-ray crystallographic studies have not been able to unambiguously differentiate between the two species. Here we use a different approach. We report a neutron crystal structure of the ferryl intermediate in Compound II of a heme peroxidase; the structure allows the protonation states of the ferryl heme to be directly observed. This, together with pre-steady state kinetic analyses, electron paramagnetic resonance spectroscopy and single crystal X-ray fluorescence, identifies a Fe(IV)-OH species as the reactive intermediate. The structure establishes a precedent for the formation of Fe(IV)-OH in a peroxidase.
Organizational Affiliation:
Department of Molecular and Cell Biology and Leicester Institute of Structural and Chemical Biology, University of Leicester, Lancaster Road, Leicester LE1 9HN, UK.