Structural characterization of the thermostable Bradyrhizobium japonicumD-sorbitol dehydrogenase.
Fredslund, F., Otten, H., Gemperlein, S., Poulsen, J.C., Carius, Y., Kohring, G.W., Lo Leggio, L.(2016) Acta Crystallogr F Struct Biol Commun 72: 846-852
- PubMed: 27827356
- DOI: https://doi.org/10.1107/S2053230X16016927
- Primary Citation of Related Structures:
5JO9 - PubMed Abstract:
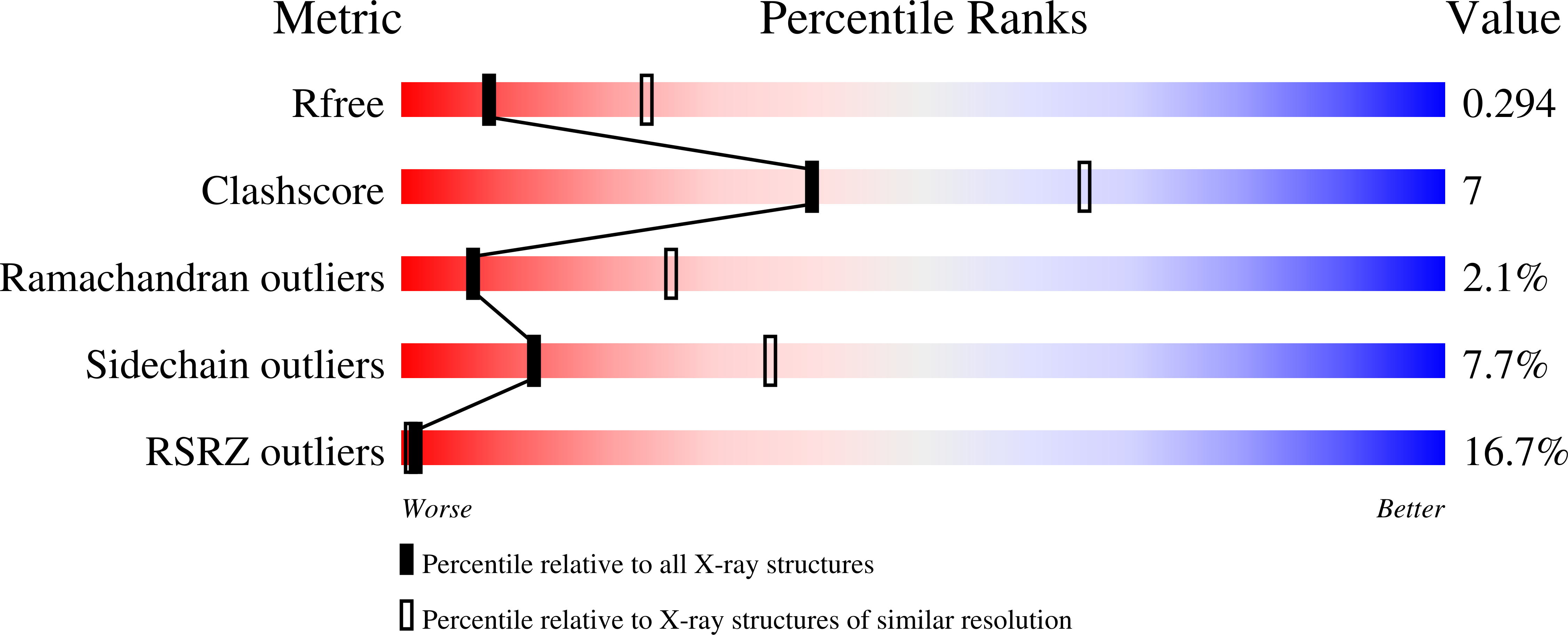
Bradyrhizobium japonicum sorbitol dehydrogenase is NADH-dependent and is active at elevated temperatures. The best substrate is D-glucitol (a synonym for D-sorbitol), although L-glucitol is also accepted, giving it particular potential in industrial applications. Crystallization led to a hexagonal crystal form, with crystals diffracting to 2.9 Å resolution. In attempts to phase the data, a molecular-replacement solution based upon PDB entry 4nbu (33% identical in sequence to the target) was found. The solution contained one molecule in the asymmetric unit, but a tetramer similar to that found in other short-chain dehydrogenases, including the search model, could be reconstructed by applying crystallographic symmetry operations. The active site contains electron density consistent with D-glucitol and phosphate, but there was not clear evidence for the binding of NADH. In a search for the features that determine the thermostability of the enzyme, the T m for the orthologue from Rhodobacter sphaeroides, for which the structure was already known, was also determined, and this enzyme proved to be considerably less thermostable. A continuous β-sheet is formed between two monomers in the tetramer of the B. japonicum enzyme, a feature not generally shared by short-chain dehydrogenases, and which may contribute to thermostability, as may an increased Pro/Gly ratio.
Organizational Affiliation:
Department of Chemistry, University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen, Denmark.