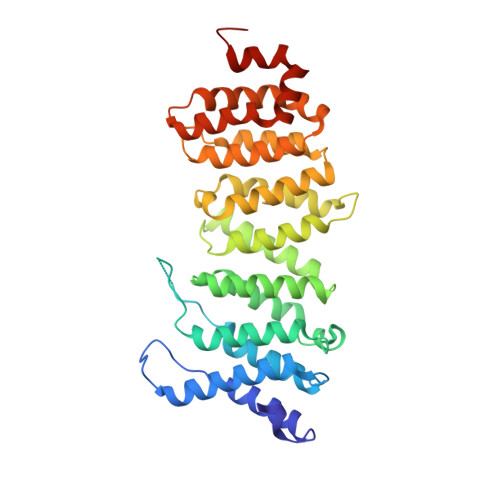
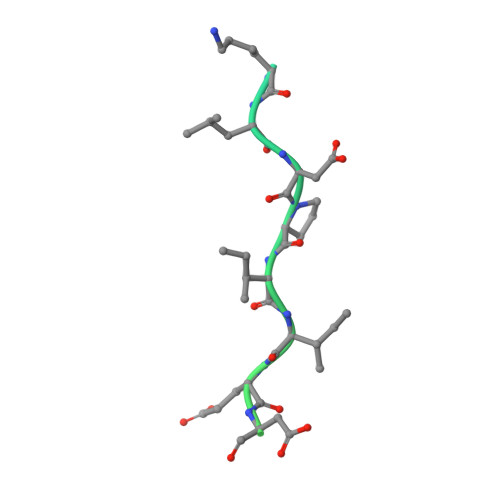
Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization.
Wang, J., Wang, Z., Yu, T., Yang, H., Virshup, D.M., Kops, G.J., Lee, S.H., Zhou, W., Li, X., Xu, W., Rao, Z.(2016) Protein Cell 7: 516-526
- PubMed: 27350047
- DOI: https://doi.org/10.1007/s13238-016-0283-4
- Primary Citation of Related Structures:
5JJA - PubMed Abstract:
Protein phosphatase 2A (PP2A) accounts for the majority of total Ser/Thr phosphatase activities in most cell types and regulates many biological processes. PP2A holoenzymes contain a scaffold A subunit, a catalytic C subunit, and one of the regulatory/targeting B subunits. How the B subunit controls PP2A localization and substrate specificity, which is a crucial aspect of PP2A regulation, remains poorly understood. The kinetochore is a critical site for PP2A functioning, where PP2A orchestrates chromosome segregation through its interactions with BubR1. The PP2A-BubR1 interaction plays important roles in both spindle checkpoint silencing and stable microtubule-kinetochore attachment. Here we present the crystal structure of a PP2A B56-BubR1 complex, which demonstrates that a conserved BubR1 LxxIxE motif binds to the concave side of the B56 pseudo-HEAT repeats. The BubR1 motif binds to a groove formed between B56 HEAT repeats 3 and 4, which is quite distant from the B56 binding surface for PP2A catalytic C subunit and thus is unlikely to affect PP2A activity. In addition, the BubR1 binding site on B56 is far from the B56 binding site of shugoshin, another kinetochore PP2A-binding protein, and thus BubR1 and shugoshin can potentially interact with PP2A-B56 simultaneously. Our structural and biochemical analysis indicates that other proteins with the LxxIxE motif may also bind to the same PP2A B56 surface. Thus, our structure of the PP2A B56-BubR1 complex provides important insights into how the B56 subunit directs the recruitment of PP2A to specific targets.
Organizational Affiliation:
College of Life Sciences, Nankai University, Tianjin, 30071, China.