Tumor Targeting with Novel 6-Substituted Pyrrolo [2,3-d] Pyrimidine Antifolates with Heteroatom Bridge Substitutions via Cellular Uptake by Folate Receptor alpha and the Proton-Coupled Folate Transporter and Inhibition of de Novo Purine Nucleotide Biosynthesis.
Golani, L.K., Wallace-Povirk, A., Deis, S.M., Wong, J., Ke, J., Gu, X., Raghavan, S., Wilson, M.R., Li, X., Polin, L., de Waal, P.W., White, K., Kushner, J., O'Connor, C., Hou, Z., Xu, H.E., Melcher, K., Dann, C.E., Matherly, L.H., Gangjee, A.(2016) J Med Chem 59: 7856-7876
- PubMed: 27458733
- DOI: https://doi.org/10.1021/acs.jmedchem.6b00594
- Primary Citation of Related Structures:
5IZQ, 5J9F - PubMed Abstract:
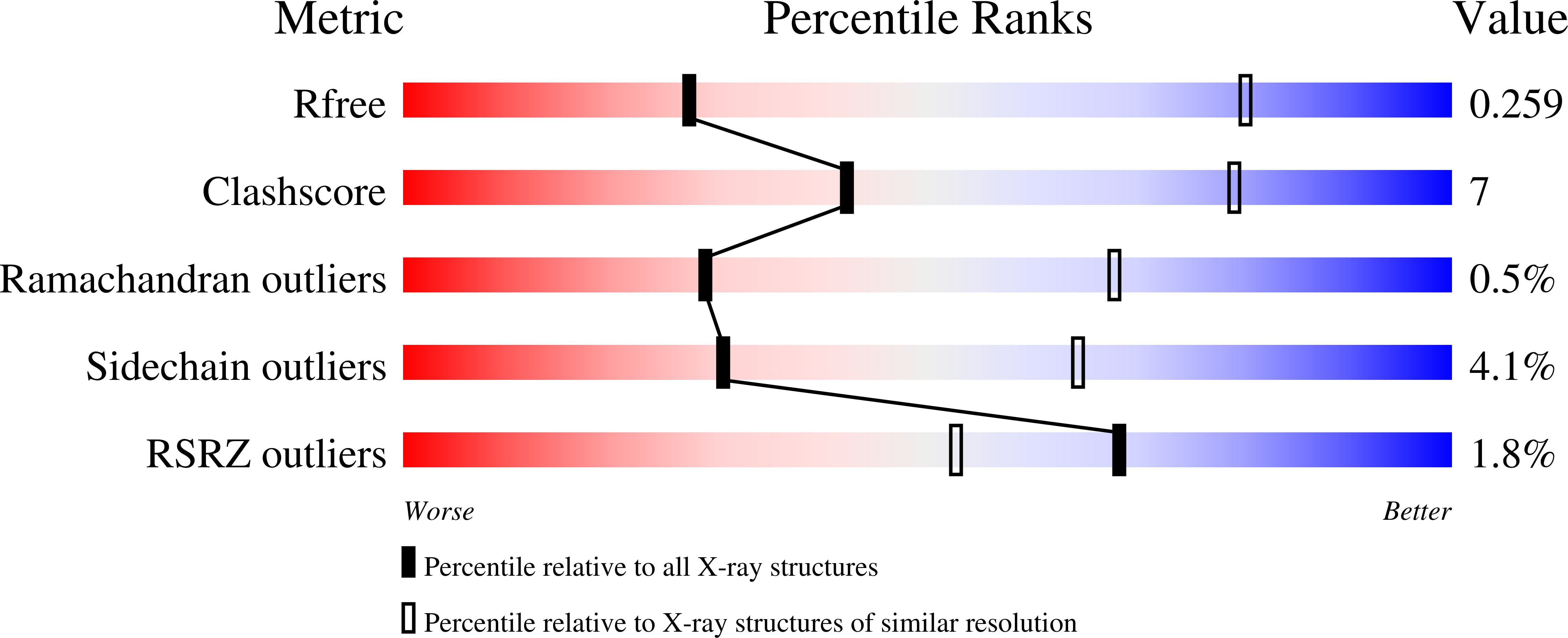
Targeted antifolates with heteroatom replacements of the carbon vicinal to the phenyl ring in 1 by N (4), O (8), or S (9), or with N-substituted formyl (5), acetyl (6), or trifluoroacetyl (7) moieties, were synthesized and tested for selective cellular uptake by folate receptor (FR) α and β or the proton-coupled folate transporter. Results show increased in vitro antiproliferative activity toward engineered Chinese hamster ovary cells expressing FRs by 4-9 over the CH2 analogue 1. Compounds 4-9 inhibited de novo purine biosynthesis and glycinamide ribonucleotide formyltransferase (GARFTase). X-ray crystal structures for 4 with FRα and GARFTase showed that the bound conformations of 4 required flexibility for attachment to both FRα and GARFTase. In mice bearing IGROV1 ovarian tumor xenografts, 4 was highly efficacious. Our results establish that heteroatom substitutions in the 3-atom bridge region of 6-substituted pyrrolo[2,3-d]pyrimidines related to 1 provide targeted antifolates that warrant further evaluation as anticancer agents.
Organizational Affiliation:
Division of Medicinal Chemistry, Graduate School of Pharmaceutical Sciences, Duquesne University , 600 Forbes Avenue, Pittsburgh, Pennsylvania 15282, United States.