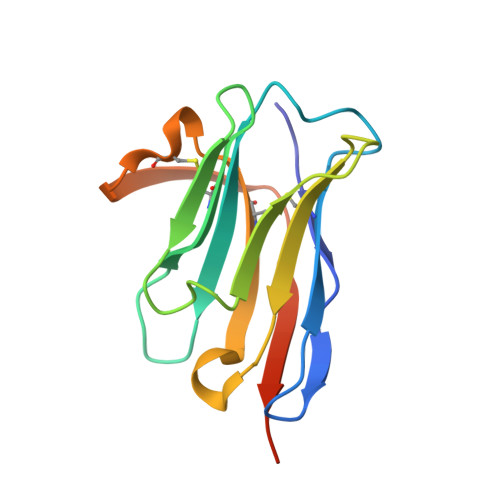
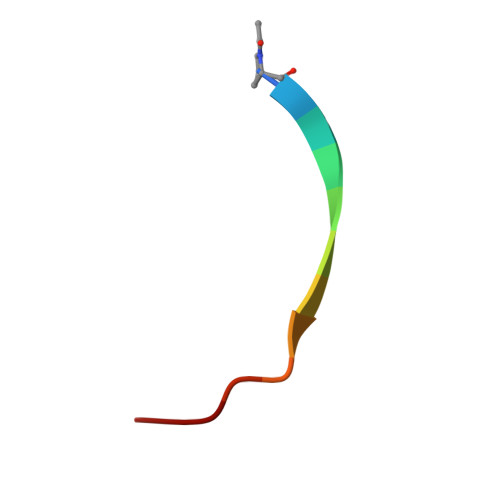
Peptides in headlock - a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy.
Braun, M.B., Traenkle, B., Koch, P.A., Emele, F., Weiss, F., Poetz, O., Stehle, T., Rothbauer, U.(2016) Sci Rep 6: 19211
- PubMed: 26791954
- DOI: https://doi.org/10.1038/srep19211
- Primary Citation of Related Structures:
5IVN, 5IVO - PubMed Abstract:
Nanobodies are highly valuable tools for numerous bioanalytical and biotechnical applications. Here, we report the characterization of a nanobody that binds a short peptide epitope with extraordinary affinity. Structural analysis reveals an unusual binding mode where the extended peptide becomes part of a β-sheet structure in the nanobody. This interaction relies on sequence-independent backbone interactions augmented by a small number of specificity-determining side chain contacts. Once bound, the peptide is fastened by two nanobody side chains that clamp it in a headlock fashion. Exploiting this unusual binding mode, we generated a novel nanobody-derived capture and detection system. Matrix-coupled nanobody enables the fast and efficient isolation of epitope-tagged proteins from prokaryotic and eukaryotic expression systems. Additionally, the fluorescently labeled nanobody visualizes subcellular structures in different cellular compartments. The high-affinity-binding and modifiable peptide tag of this system renders it a versatile and robust tool to combine biochemical analysis with microscopic studies.
Organizational Affiliation:
Interfaculty Institute of Biochemistry, Eberhard-Karls University Tuebingen, Germany.