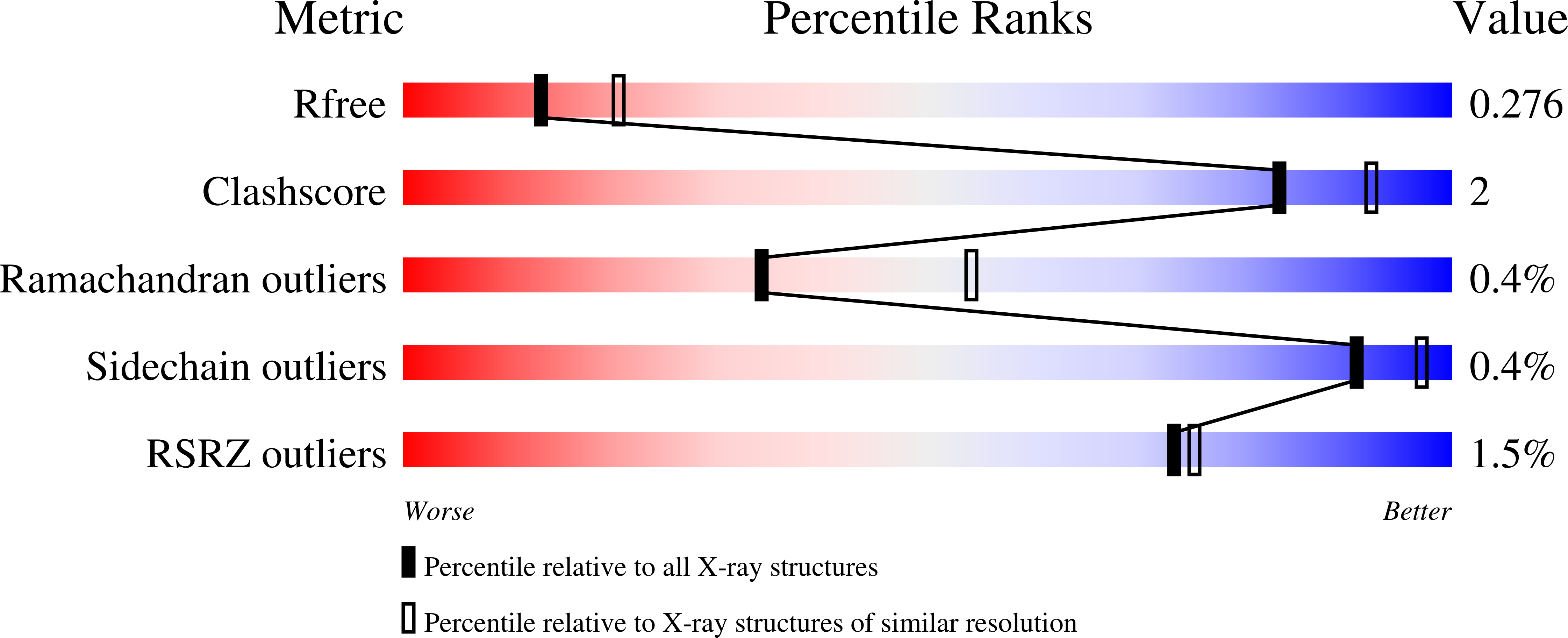
Structural basis for halogenation by iron- and 2-oxo-glutarate-dependent enzyme WelO5.
Mitchell, A.J., Zhu, Q., Maggiolo, A.O., Ananth, N.R., Hillwig, M.L., Liu, X., Boal, A.K.(2016) Nat Chem Biol 12: 636-640
- PubMed: 27348090
- DOI: https://doi.org/10.1038/nchembio.2112
- Primary Citation of Related Structures:
5IQS, 5IQT, 5IQU, 5IQV - PubMed Abstract:
A 2.4-Å-resolution X-ray crystal structure of the carrier-protein-independent halogenase WelO5 in complex with its welwitindolinone precursor substrate, 12-epi-fischerindole U, reveals that the C13 chlorination target is proximal to the anticipated site of the oxo group in a presumptive cis-halo-oxo-iron(IV) (haloferryl) intermediate. Prior study of related halogenases forecasts substrate hydroxylation in this active-site configuration, but X-ray crystallographic verification of C13 halogenation in single crystals mandates that ligand dynamics must reposition the oxygen ligand to enable the observed outcome. S189A WelO5 produces a mixture of halogenation and hydroxylation products, showing that an outer-sphere hydrogen-bonding group orchestrates ligand movements to achieve a configuration that promotes halogen transfer.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania, USA.