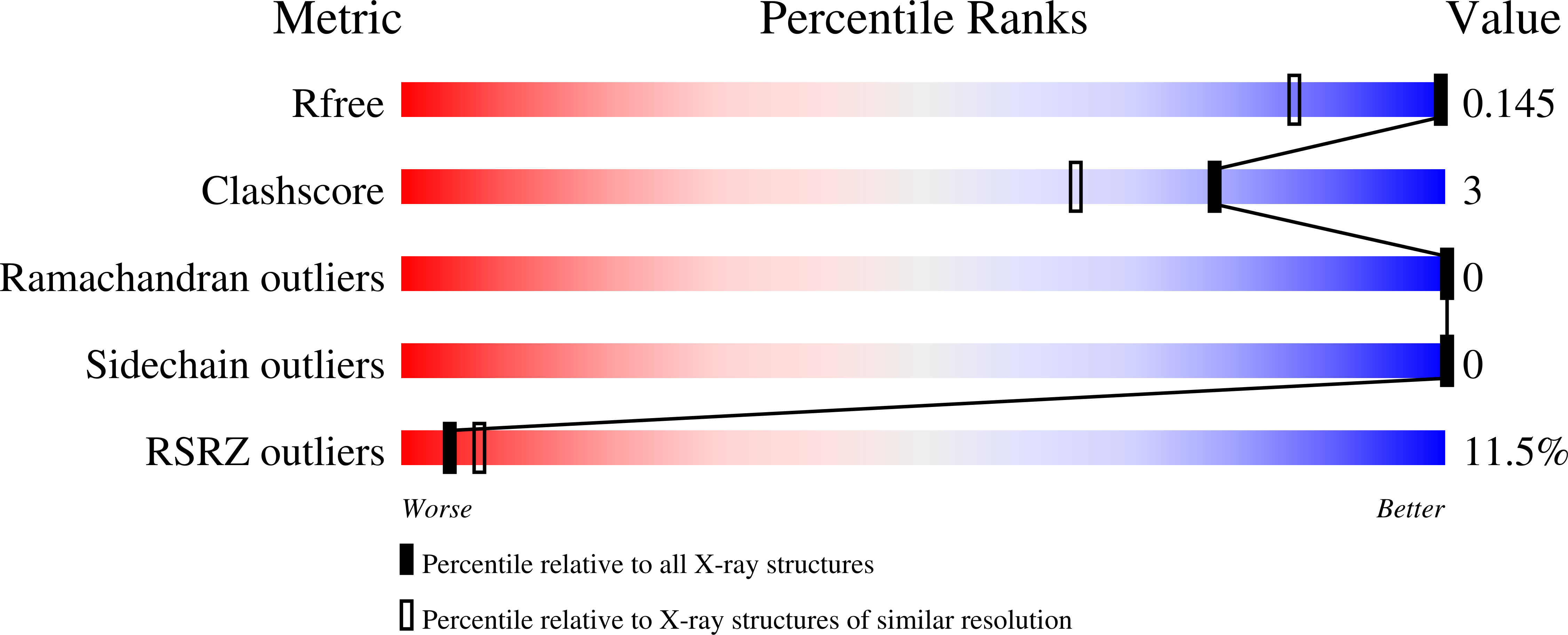
Peroxiredoxin Catalysis at Atomic Resolution.
Perkins, A., Parsonage, D., Nelson, K.J., Ogba, O.M., Cheong, P.H., Poole, L.B., Karplus, P.A.(2016) Structure 24: 1668-1678
- PubMed: 27594682
- DOI: https://doi.org/10.1016/j.str.2016.07.012
- Primary Citation of Related Structures:
5IIZ, 5IM9, 5IMA, 5IMC, 5IMD, 5IMF, 5IMV, 5IMZ, 5INY, 5IO0, 5IO2, 5IOW, 5IOX, 5IPG, 5IPH - PubMed Abstract:
Peroxiredoxins (Prxs) are ubiquitous cysteine-based peroxidases that guard cells against oxidative damage, are virulence factors for pathogens, and are involved in eukaryotic redox regulatory pathways. We have analyzed catalytically active crystals to capture atomic resolution snapshots of a PrxQ subfamily enzyme (from Xanthomonas campestris) proceeding through thiolate, sulfenate, and sulfinate species. These analyses provide structures of unprecedented accuracy for seeding theoretical studies, and reveal conformational intermediates giving insight into the reaction pathway. Based on a highly non-standard geometry seen for the sulfenate intermediate, we infer that the sulfenate formation itself can strongly promote local unfolding of the active site to enhance productive catalysis. Further, these structures reveal that preventing local unfolding, in this case via crystal contacts, results in facile hyperoxidative inactivation even for Prxs normally resistant to such inactivation. This supports previous proposals that conformation-specific inhibitors may be useful for achieving selective inhibition of Prxs that are drug targets.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Oregon State University, Corvallis, OR 97331, USA.