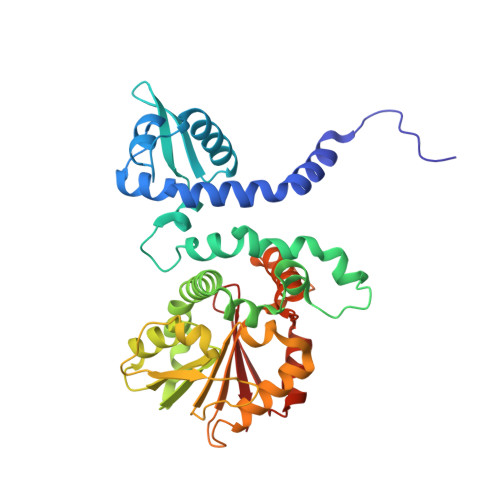
Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Robin, A.Y., Giustini, C., Graindorge, M., Matringe, M., Dumas, R.(2016) Plant J 87: 641-653
- PubMed: 27232113
- DOI: https://doi.org/10.1111/tpj.13225
- Primary Citation of Related Structures:
5ICC, 5ICE, 5ICF, 5ICG - PubMed Abstract:
Growing pharmaceutical interest in benzylisoquinoline alkaloids (BIA) coupled with their chemical complexity make metabolic engineering of microbes to create alternative platforms of production an increasingly attractive proposition. However, precise knowledge of rate-limiting enzymes and negative feedback inhibition by end-products of BIA metabolism is of paramount importance for this emerging field of synthetic biology. In this work we report the structural characterization of (S)-norcoclaurine-6-O-methyltransferase (6OMT), a key rate-limiting step enzyme involved in the synthesis of reticuline, the final intermediate to be shared between the different end-products of BIA metabolism, such as morphine, papaverine, berberine and sanguinarine. Four different crystal structures of the enzyme from Thalictrum flavum (Tf 6OMT) were solved: the apoenzyme, the complex with S-adenosyl-l-homocysteine (SAH), the complexe with SAH and the substrate and the complex with SAH and a feedback inhibitor, sanguinarine. The Tf 6OMT structural study provides a molecular understanding of its substrate specificity, active site structure and reaction mechanism. This study also clarifies the inhibition of Tf 6OMT by previously suggested feedback inhibitors. It reveals its high and time-dependent sensitivity toward sanguinarine.
Organizational Affiliation:
Laboratoire de Physiologie Cellulaire et Végétale, Université Grenoble Alpes, CNRS UMR5168, CEA/DRF/BIG, INRA UMR 1417, 17, Avenue des Martyrs, 38054 Grenoble, France.