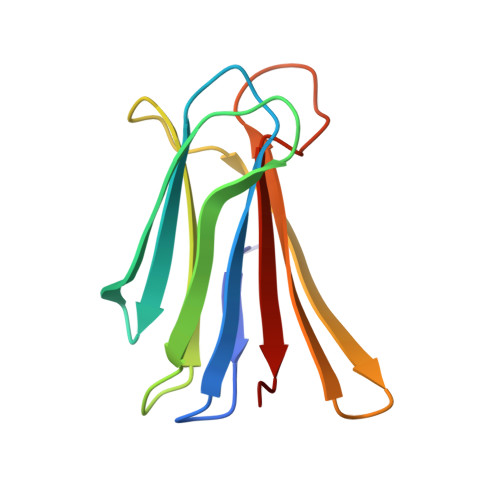
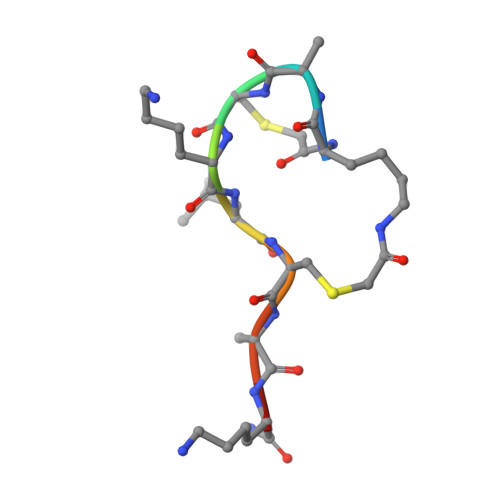
Chemical space guided discovery of antimicrobial bridged bicyclic peptides against Pseudomonas aeruginosa and its biofilms.
Di Bonaventura, I., Jin, X., Visini, R., Probst, D., Javor, S., Gan, B.H., Michaud, G., Natalello, A., Doglia, S.M., Kohler, T., van Delden, C., Stocker, A., Darbre, T., Reymond, J.L.(2017) Chem Sci 8: 6784-6798
- PubMed: 29147502
- DOI: https://doi.org/10.1039/c7sc01314k
- Primary Citation of Related Structures:
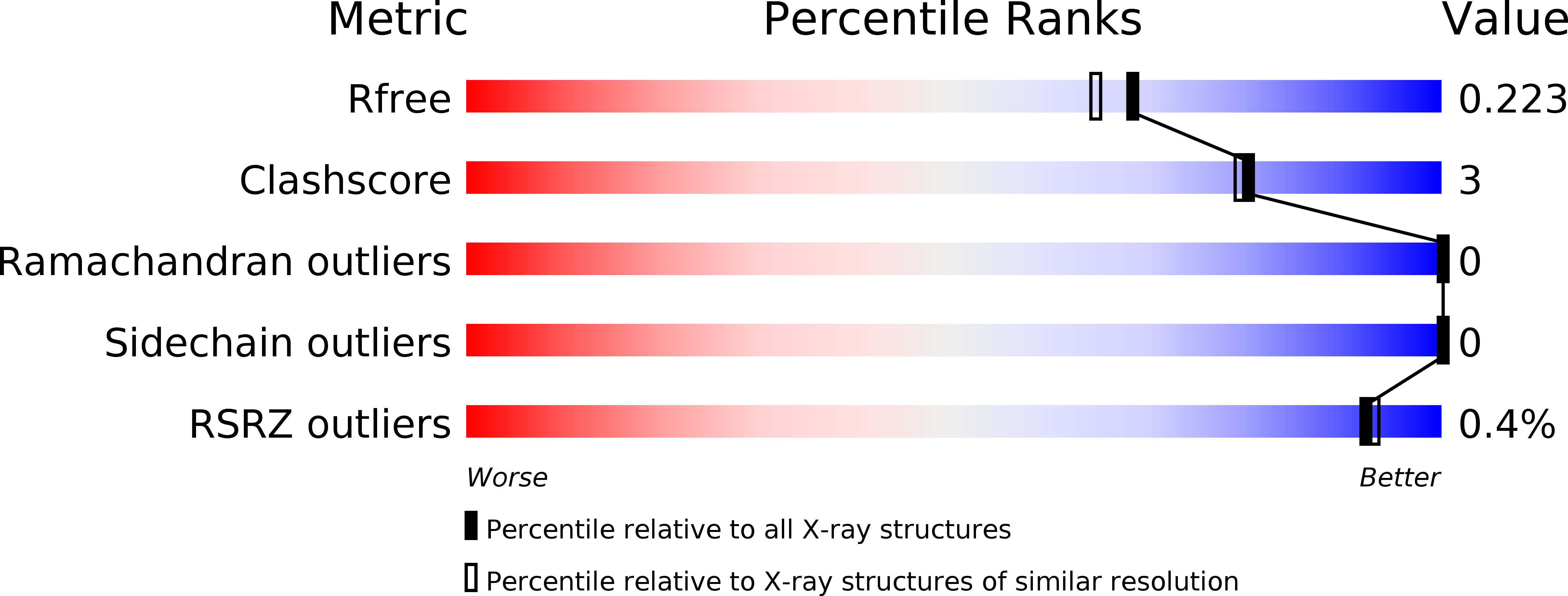
5I8M, 5I8X, 5NGQ - PubMed Abstract:
Herein we report the discovery of antimicrobial bridged bicyclic peptides (AMBPs) active against Pseudomonas aeruginosa , a highly problematic Gram negative bacterium in the hospital environment. Two of these AMBPs show strong biofilm inhibition and dispersal activity and enhance the activity of polymyxin, currently a last resort antibiotic against which resistance is emerging. To discover our AMBPs we used the concept of chemical space, which is well known in the area of small molecule drug discovery, to define a small number of test compounds for synthesis and experimental evaluation. Our chemical space was calculated using 2DP, a new topological shape and pharmacophore fingerprint for peptides. This method provides a general strategy to search for bioactive peptides with unusual topologies and expand the structural diversity of peptide-based drugs.
Organizational Affiliation:
Department of Chemistry and Biochemistry , University of Bern , Freiestrasse 3 , 3012 Bern , Switzerland . Email: jean-louis.reymond@dcb.unibe.ch.