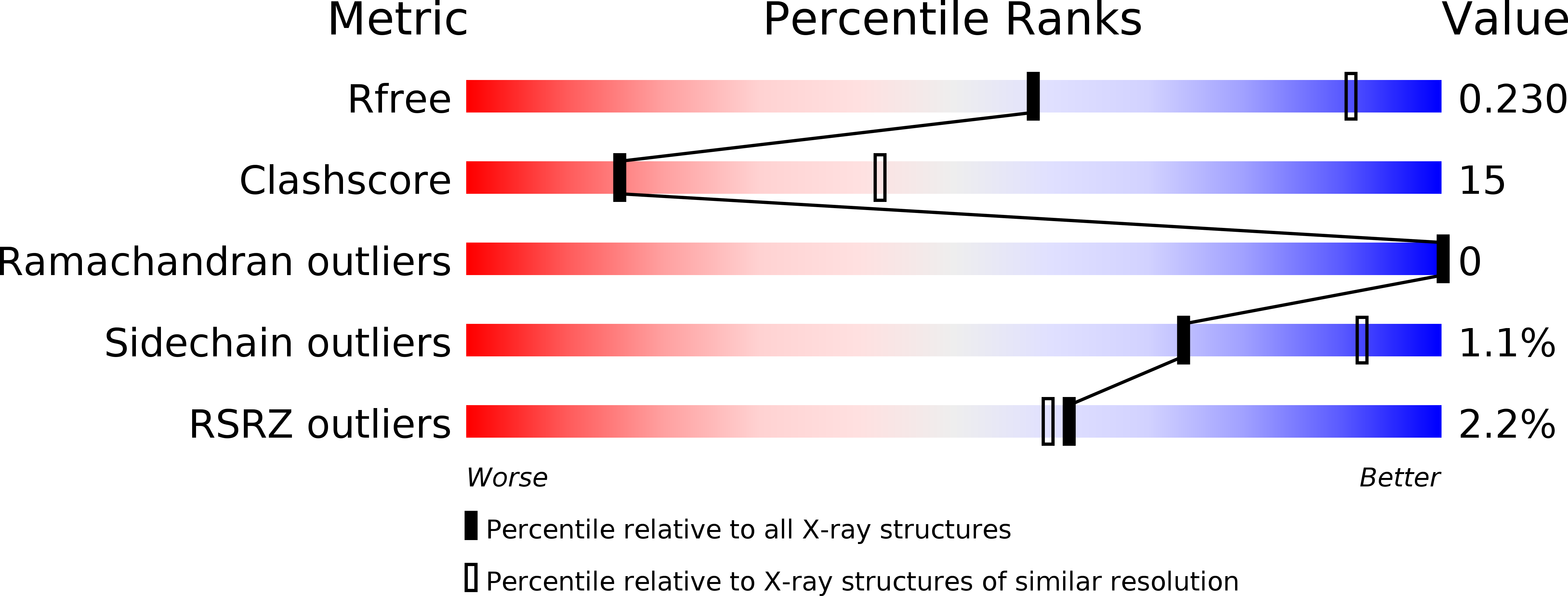
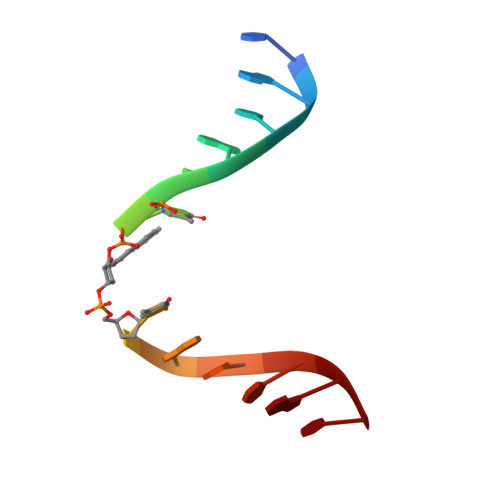
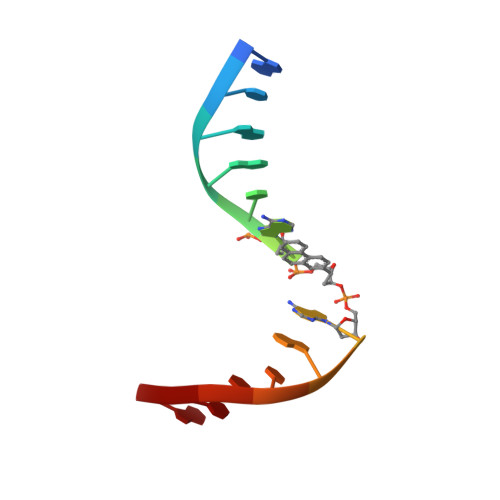
X-ray structure of a lectin-bound DNA duplex containing an unnatural phenanthrenyl pair.
Roethlisberger, P., Istrate, A., Marcaida Lopez, M.J., Visini, R., Stocker, A., Reymond, J.L., Leumann, C.J.(2016) Chem Commun (Camb) 52: 4749-4752
- PubMed: 26898721
- DOI: https://doi.org/10.1039/c6cc00374e
- Primary Citation of Related Structures:
5HCH - PubMed Abstract:
DNA duplexes containing unnatural base-pair surrogates are attractive biomolecular nanomaterials with potentially beneficial photophysical or electronic properties. Herein we report the first X-ray structure of a duplex containing a phen-pair in the center of the double helix in a zipper like stacking arrangement.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Bern, Freiestrasse 3, 3012 Bern, Switzerland. christian.leumann@dcb.unibe.ch.