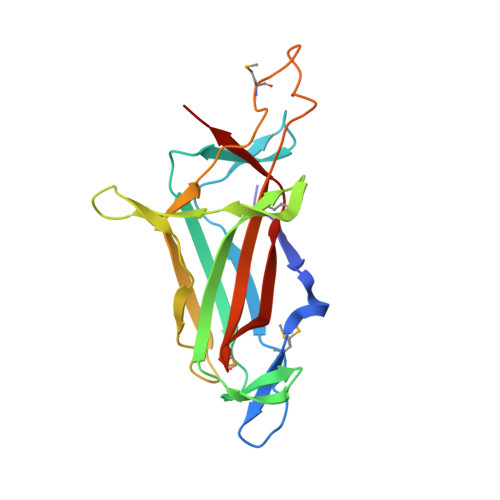
Crystal structure of the DNA-binding domain of Myelin-gene Regulatory Factor.
Zhen, X., Li, B., Hu, F., Yan, S., Meloni, G., Li, H., Shi, N.(2017) Sci Rep 7: 3696-3696
- PubMed: 28623291
- DOI: https://doi.org/10.1038/s41598-017-03768-9
- Primary Citation of Related Structures:
5H5P - PubMed Abstract:
Myelin-gene Regulatory Factor (MyRF) is one of the master transcription factors controlling myelin formation and development in oligodendrocytes which is crucial for the powerful brain functions. The N-terminal of MyRF, which contains a proline-rich region and a DNA binding domain (DBD), is auto-cleaved from the ER membrane, and then enters the nucleus to participate in transcription regulation of the myelin genes. Here we report the crystal structure of MyRF DBD. It shows an Ig-fold like architecture which consists of two antiparallel β-sheets with 7 main strands, packing against each other, forming a β-sandwich. Compared to its homolog, Ndt80, MyRF has a smaller and less complex DBD lacking the helices and the big loops outside the core. Structural alignment reveals that MyRF DBD possess less interaction sites with DNA than Ndt80 and may bind only at the major groove of DNA. Moreover, the structure reveals a trimeric assembly, agreeing with the previous report that MyRF DBD functions as a trimer. The mutant that we designed based on the structure disturbed trimer formation, but didn't affect the auto-cleavage reaction. It demonstrates that the activation of self-cleavage reaction of MyRF is independent of the presence of its N-terminal DBD homotrimer. The structure reported here will help to understand the molecular mechanism underlying the important roles of MyRF in myelin formation and development.
Organizational Affiliation:
State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou, 350002, China.