Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Jin, W., Wachino, J., Yamaguchi, Y., Kimura, K., Kumar, A., Yamada, M., Morinaka, A., Sakamaki, Y., Yonezawa, M., Kurosaki, H., Arakawa, Y.(2017) Antimicrob Agents Chemother 61
- PubMed: 28739781
- DOI: https://doi.org/10.1128/AAC.00501-17
- Primary Citation of Related Structures:
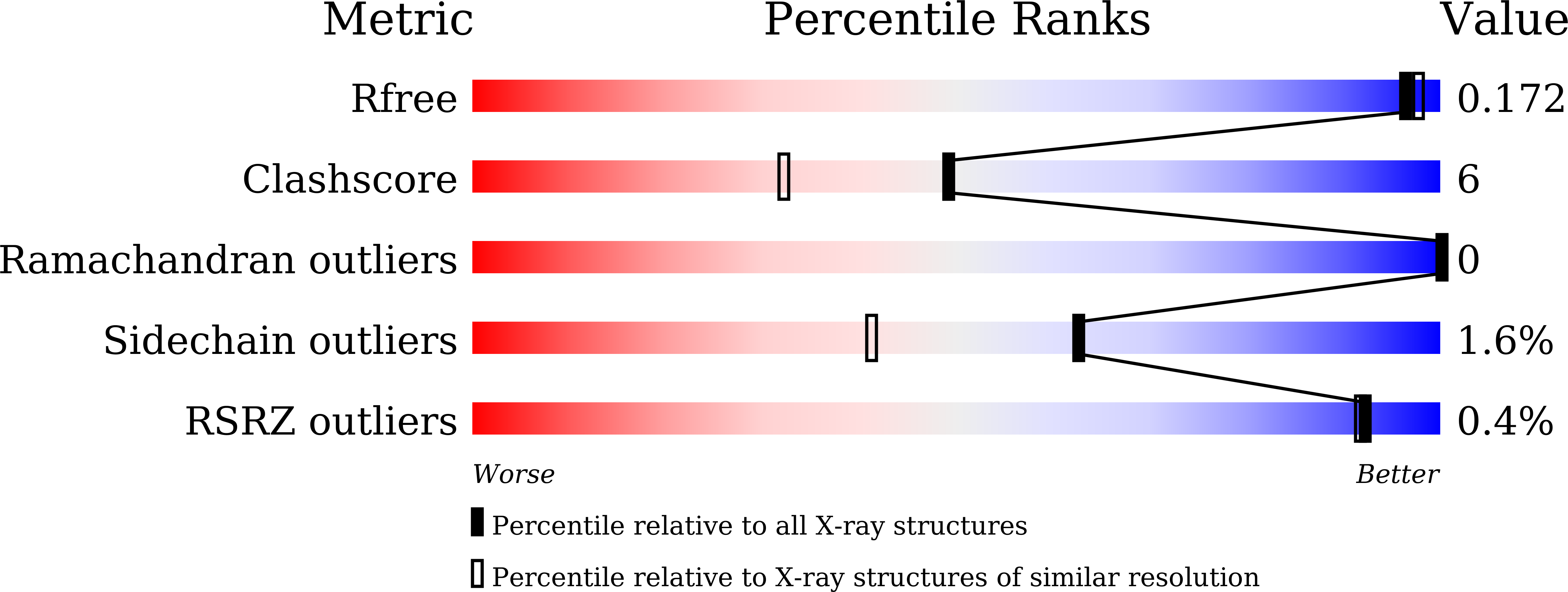
5GS8, 5GWA, 5X5G - PubMed Abstract:
The development of effective inhibitors that block extended-spectrum β-lactamases (ESBLs) and restore the action of β-lactams represents an effective strategy against ESBL-producing Enterobacteriaceae We evaluated the inhibitory effects of the diazabicyclooctanes avibactam and OP0595 against TLA-3, an ESBL that we identified previously. Avibactam and OP0595 inhibited TLA-3 with apparent inhibitor constants ( K i app ) of 1.71 ± 0.10 and 1.49 ± 0.05 μM, respectively, and could restore susceptibility to cephalosporins in the TLA-3-producing Escherichia coli strain. The value of the second-order acylation rate constant ( k 2 / K , where k 2 is the acylation rate constant and K is the equilibrium constant) of avibactam [(3.25 ± 0.03) × 10 3 M -1 · s -1 ] was closer to that of class C and D β-lactamases ( k 2 / K , <10 4 M -1 · s -1 ) than that of class A β-lactamases ( k 2 / K , >10 4 M -1 · s -1 ). In addition, we determined the structure of TLA-3 and that of TLA-3 complexed with avibactam or OP0595 at resolutions of 1.6, 1.6, and 2.0 Å, respectively. TLA-3 contains an inverted Ω loop and an extended loop between the β5 and β6 strands (insertion after Ser237), which appear only in PER-type class A β-lactamases. These structures might favor the accommodation of cephalosporins harboring bulky R1 side chains. TLA-3 presented a high catalytic efficiency ( k cat / K m ) against cephalosporins, including cephalothin, cefuroxime, and cefotaxime. Avibactam and OP0595 bound covalently to TLA-3 via the Ser70 residue and made contacts with residues Ser130, Thr235, and Ser237, which are conserved in ESBLs. Additionally, the sulfate group of the inhibitors formed polar contacts with amino acid residues in a positively charged pocket of TLA-3. Our findings provide a structural template for designing improved diazabicyclooctane-based inhibitors that are effective against ESBL-producing Enterobacteriaceae .
Organizational Affiliation:
Department of Bacteriology, Nagoya University Graduate School of Medicine, Nagoya, Aichi, Japan.