Discovery and Characterization of a Thermostable and Highly Halotolerant Gh5 Cellulase from an Icelandic Hot Spring Isolate.
Zarafeta, D., Kissas, D., Sayer, C., Gudbergsdottir, S.R., Ladoukakis, E., Isupov, M.N., Chatziioannou, A., Peng, X., Littlechild, J.A., Skretas, G., Kolisis, F.N.(2016) PLoS One 11: 46454
- PubMed: 26741138
- DOI: https://doi.org/10.1371/journal.pone.0146454
- Primary Citation of Related Structures:
5FIP - PubMed Abstract:
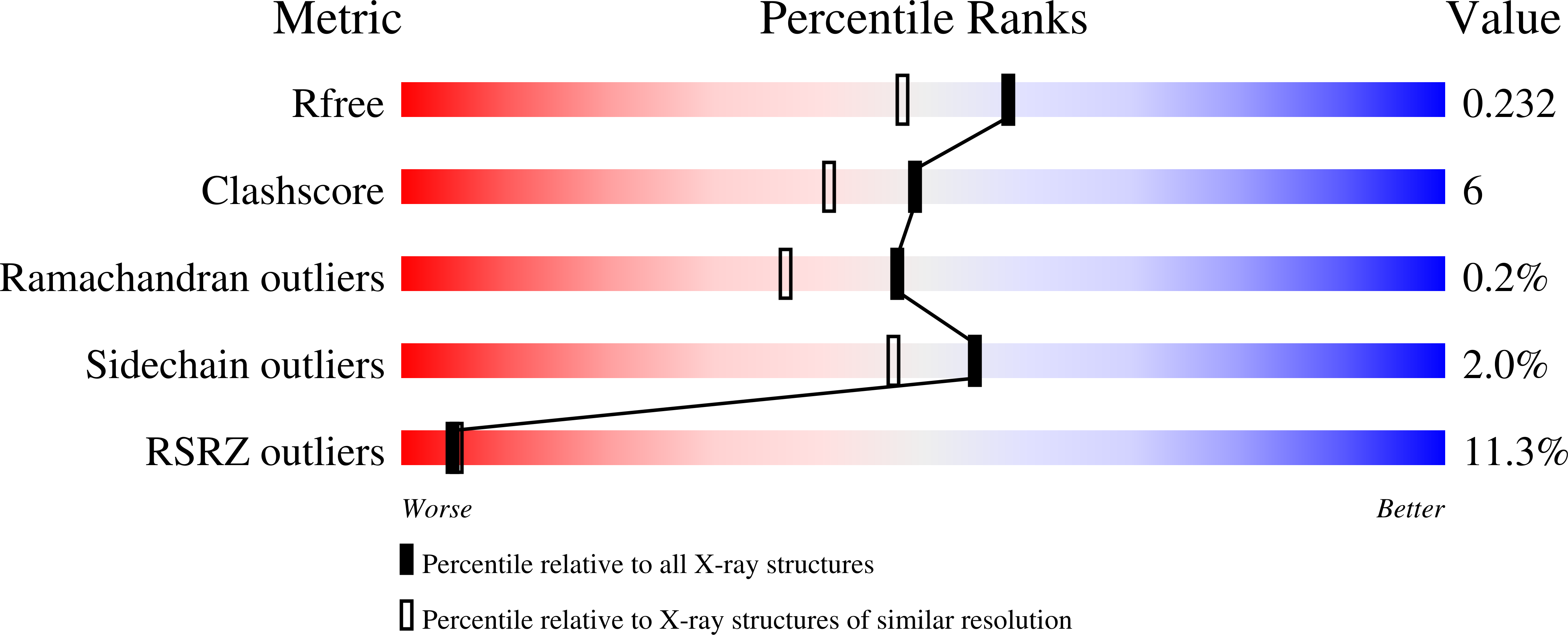
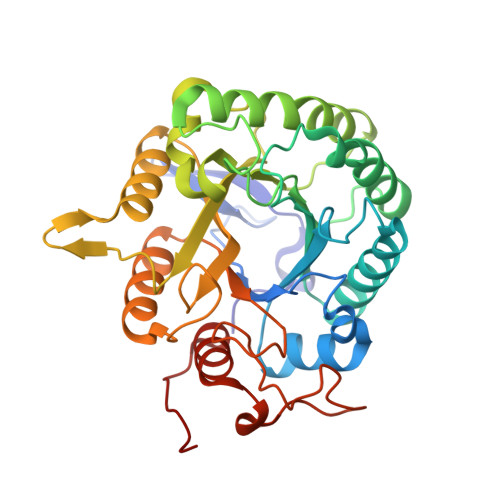
With the ultimate goal of identifying robust cellulases for industrial biocatalytic conversions, we have isolated and characterized a new thermostable and very halotolerant GH5 cellulase. This new enzyme, termed CelDZ1, was identified by bioinformatic analysis from the genome of a polysaccharide-enrichment culture isolate, initiated from material collected from an Icelandic hot spring. Biochemical characterization of CelDZ1 revealed that it is a glycoside hydrolase with optimal activity at 70°C and pH 5.0 that exhibits good thermostability, high halotolerance at near-saturating salt concentrations, and resistance towards metal ions and other denaturing agents. X-ray crystallography of the new enzyme showed that CelDZ1 is the first reported cellulase structure that lacks the defined sugar-binding 2 subsite and revealed structural features which provide potential explanations of its biochemical characteristics.
Organizational Affiliation:
Institute of Biology, Medicinal Chemistry & Biotechnology, National Hellenic Research Foundation, Athens, Greece.