Structure and function of the yeast listerin (Ltn1) conserved N-terminal domain in binding to stalled 60S ribosomal subunits.
Doamekpor, S.K., Lee, J.W., Hepowit, N.L., Wu, C., Charenton, C., Leonard, M., Bengtson, M.H., Rajashankar, K.R., Sachs, M.S., Lima, C.D., Joazeiro, C.A.(2016) Proc Natl Acad Sci U S A 113: E4151-E4160
- PubMed: 27385828
- DOI: https://doi.org/10.1073/pnas.1605951113
- Primary Citation of Related Structures:
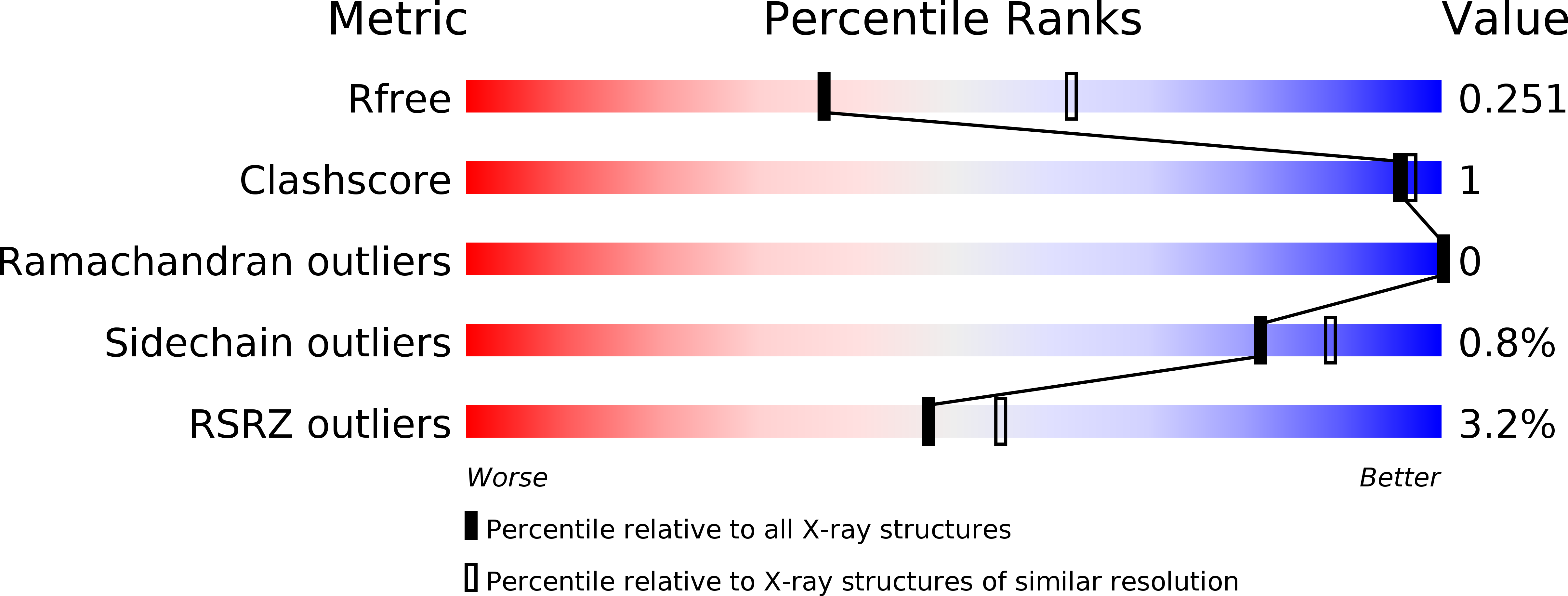
5FG0, 5FG1 - PubMed Abstract:
The Ltn1 E3 ligase (listerin in mammals) has emerged as a paradigm for understanding ribosome-associated ubiquitylation. Ltn1 binds to 60S ribosomal subunits to ubiquitylate nascent polypeptides that become stalled during synthesis; among Ltn1's substrates are aberrant products of mRNA lacking stop codons [nonstop translation products (NSPs)]. Here, we report the reconstitution of NSP ubiquitylation in Neurospora crassa cell extracts. Upon translation in vitro, ribosome-stalled NSPs were ubiquitylated in an Ltn1-dependent manner, while still ribosome-associated. Furthermore, we provide biochemical evidence that the conserved N-terminal domain (NTD) plays a significant role in the binding of Ltn1 to 60S ribosomal subunits and that NTD mutations causing defective 60S binding also lead to defective NSP ubiquitylation, without affecting Ltn1's intrinsic E3 ligase activity. Finally, we report the crystal structure of the Ltn1 NTD at 2.4-Å resolution. The structure, combined with additional mutational studies, provides insight to NTD's role in binding stalled 60S subunits. Our findings show that Neurospora extracts can be used as a tool to dissect mechanisms underlying ribosome-associated protein quality control and are consistent with a model in which Ltn1 uses 60S subunits as adapters, at least in part via its NTD, to target stalled NSPs for ubiquitylation.
Organizational Affiliation:
Structural Biology Program, Sloan-Kettering Institute, New York, NY 10065;