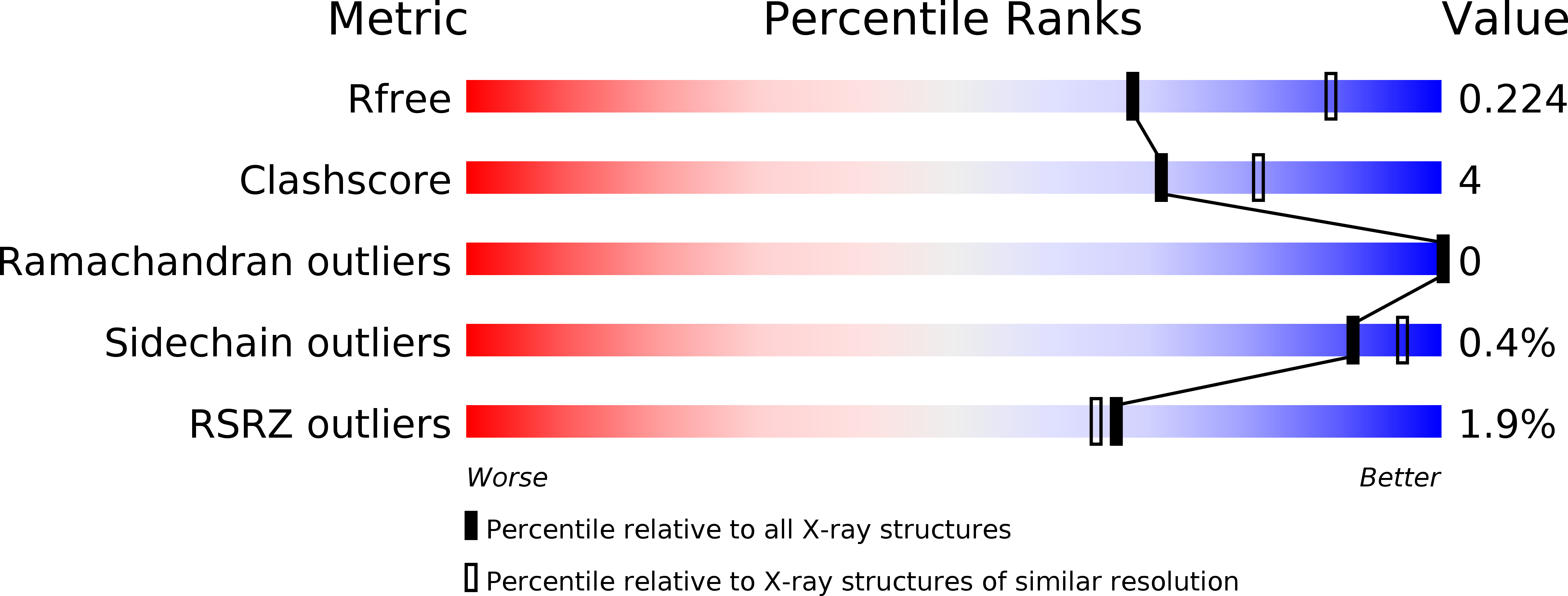
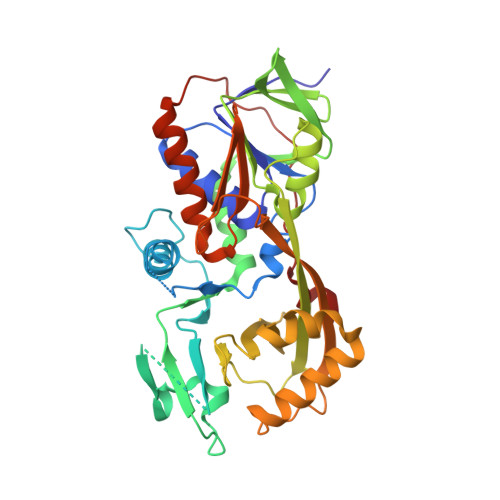
Crystal structure of the flavoenzyme PA4991 from Pseudomonas aeruginosa.
Jacewicz, A., Schnell, R., Lindqvist, Y., Schneider, G.(2016) Acta Crystallogr F Struct Biol Commun 72: 105-111
- PubMed: 26841760
- DOI: https://doi.org/10.1107/S2053230X15024437
- Primary Citation of Related Structures:
5EZ7 - PubMed Abstract:
The locus PA4991 in Pseudomonas aeruginosa encodes an open reading frame that has been identified as essential for the virulence and/or survival of this pathogenic organism in the infected host. Here, it is shown that this gene encodes a monomeric FAD-binding protein of molecular mass 42.2 kDa. The structure of PA4991 was determined by a combination of molecular replacement using a search model generated with Rosetta and phase improvement by a low-occupancy heavy-metal derivative. PA4991 belongs to the GR2 family of FAD-dependent oxidoreductases, comprising an FAD-binding domain typical of the glutathione reductase family and a second domain dominated by an eight-stranded mixed β-sheet. Most of the protein-FAD interactions are via the FAD-binding domain, but the isoalloxazine ring is located at the domain interface and interacts with residues from both domains. A comparison with the structurally related glycine oxidase and glycerol-3-phosphate dehydrogenase shows that in spite of very low amino-acid sequence identity (<18%) several active-site residues involved in substrate binding in these enzymes are conserved in PA4991. However, enzymatic assays show that PA4991 does not display amino-acid oxidase or glycerol-3-phosphate dehydrogenase activities, suggesting that it requires different substrates for activity.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Karolinska Institutet, S-171 77 Stockholm, Sweden.