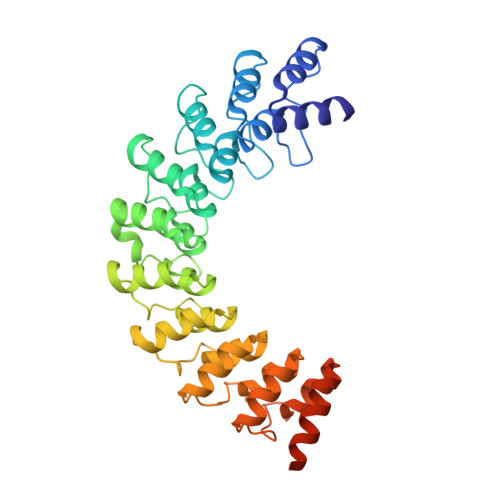
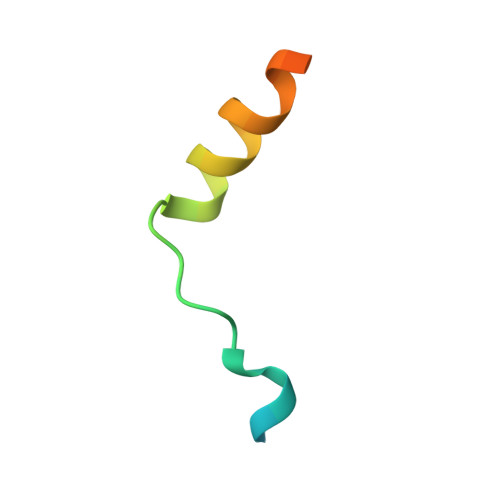
Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Liu, H., Li, J., Raval, M.H., Yao, N., Deng, X., Lu, Q., Nie, S., Feng, W., Wan, J., Yengo, C.M., Liu, W., Zhang, M.(2016) Elife 5
- PubMed: 26785147
- DOI: https://doi.org/10.7554/eLife.12856
- Primary Citation of Related Structures:
5ET0, 5ET1 - PubMed Abstract:
Class III myosins (Myo3) and actin-bundling protein Espin play critical roles in regulating the development and maintenance of stereocilia in vertebrate hair cells, and their defects cause hereditary hearing impairments. Myo3 interacts with Espin1 through its tail homology I motif (THDI), however it is not clear how Myo3 specifically acts through Espin1 to regulate the actin bundle assembly and stabilization. Here we discover that Myo3 THDI contains a pair of repeat sequences capable of independently and strongly binding to the ankyrin repeats of Espin1, revealing an unexpected Myo3-mediated cross-linking mechanism of Espin1. The structures of Myo3 in complex with Espin1 not only elucidate the mechanism of the binding, but also reveal a Myo3-induced release of Espin1 auto-inhibition mechanism. We also provide evidence that Myo3-mediated cross-linking can further promote actin fiber bundling activity of Espin1.
Organizational Affiliation:
Shenzhen Key Laboratory for Neuronal Structural Biology, Biomedical Research Institute, Shenzhen Peking University-The Hong Kong University of Science and Technology Medical Center, Shenzhen, China.