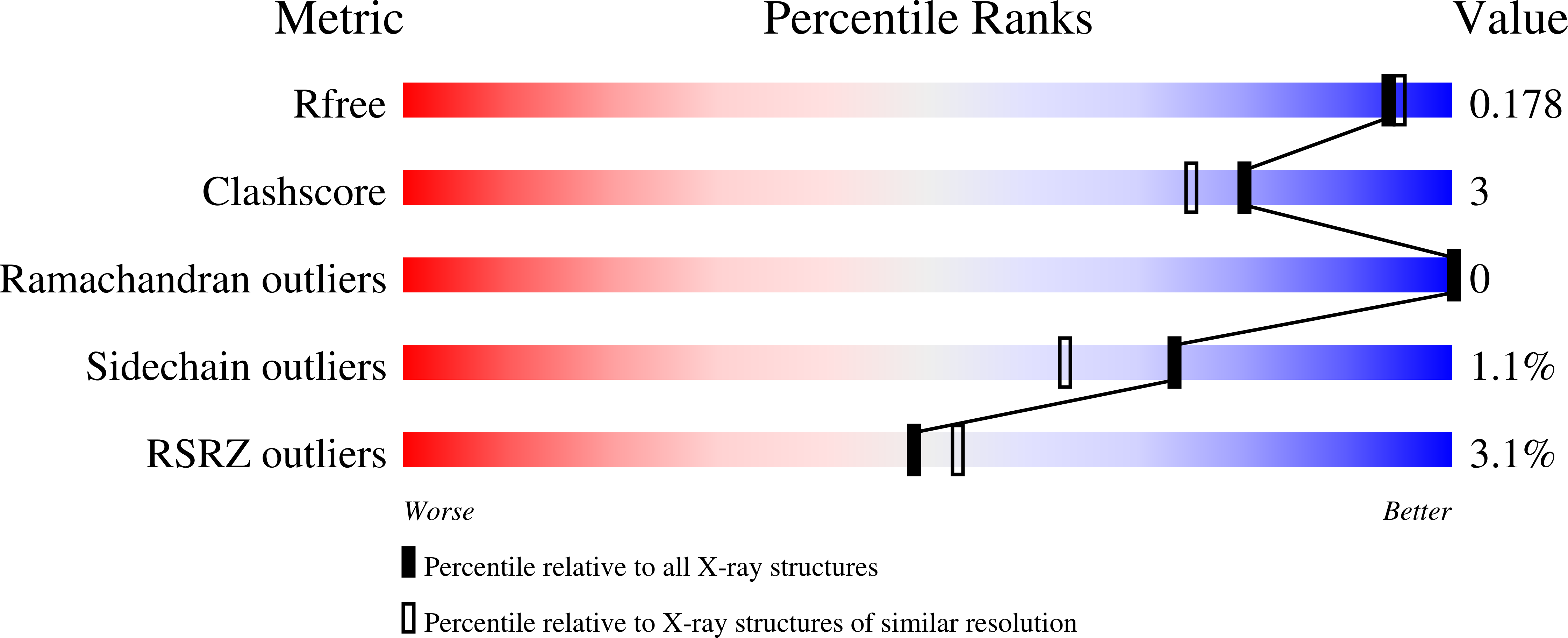
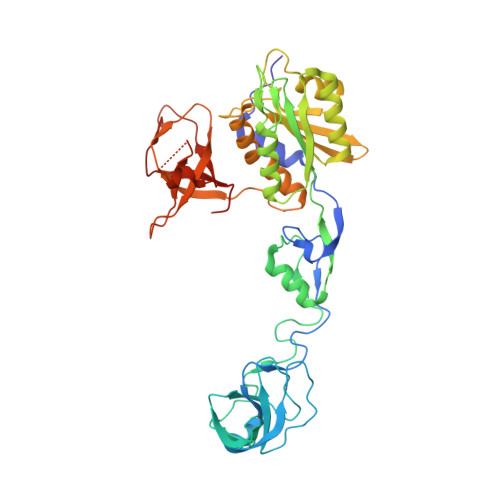
Structural Framework for Metal Incorporation during Molybdenum Cofactor Biosynthesis.
Kasaragod, V.B., Schindelin, H.(2016) Structure 24: 782-788
- PubMed: 27112598
- DOI: https://doi.org/10.1016/j.str.2016.02.023
- Primary Citation of Related Structures:
5ERQ, 5ERR, 5ERS, 5ERT, 5ERU, 5ERV - PubMed Abstract:
The molybdenum cofactor (Moco) is essential for the catalytic activity of all molybdenum-containing enzymes with the exception of nitrogenase. Moco biosynthesis follows an evolutionarily highly conserved pathway and genetic deficiencies in the corresponding human enzymes result in Moco deficiency, which manifests itself in severe neurological symptoms and death in childhood. In humans the final steps of Moco biosynthesis are catalyzed by gephyrin, specifically the penultimate adenylation of molybdopterin (MPT) by its N-terminal G domain (GephG) and the final metal incorporation by its C-terminal E domain (GephE). To better understand the poorly defined molecular framework of this final step, we determined high-resolution crystal structures of GephE in the apo state and in complex with ADP, AMP, and molybdate. Our data provide novel insights into the catalytic steps leading to final Moco maturation, namely deadenylation as well as molybdate binding and insertion.
Organizational Affiliation:
Rudolf Virchow Center for Experimental Biomedicine, Institute of Structural Biology, University of Würzburg, Josef-Schneider-Straße 2, 97080 Würzburg, Germany.