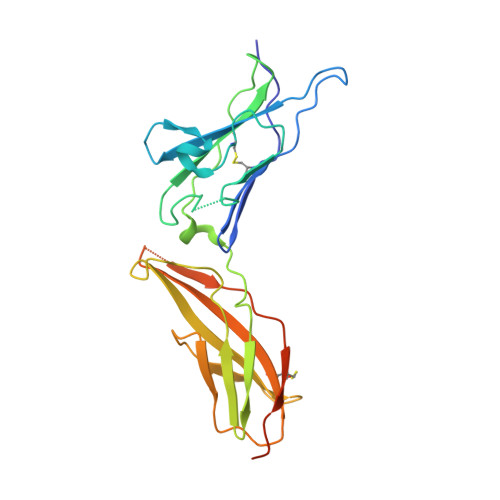
Crystal structure of human interferon-gamma receptor 2 reveals the structural basis for receptor specificity.
Mikulecky, P., Zahradnik, J., Kolenko, P., Cerny, J., Charnavets, T., Kolarova, L., Necasova, I., Pham, P.N., Schneider, B.(2016) Acta Crystallogr D Struct Biol 72: 1017-1025
- PubMed: 27599734
- DOI: https://doi.org/10.1107/S2059798316012237
- Primary Citation of Related Structures:
5EH1 - PubMed Abstract:
Interferon-γ receptor 2 is a cell-surface receptor that is required for interferon-γ signalling and therefore plays a critical immunoregulatory role in innate and adaptive immunity against viral and also bacterial and protozoal infections. A crystal structure of the extracellular part of human interferon-γ receptor 2 (IFNγR2) was solved by molecular replacement at 1.8 Å resolution. Similar to other class 2 receptors, IFNγR2 has two fibronectin type III domains. The characteristic structural features of IFNγR2 are concentrated in its N-terminal domain: an extensive π-cation motif of stacked residues KWRWRH, a NAG-W-NAG sandwich (where NAG stands for N-acetyl-D-glucosamine) and finally a helix formed by residues 78-85, which is unique among class 2 receptors. Mass spectrometry and mutational analyses showed the importance of N-linked glycosylation to the stability of the protein and confirmed the presence of two disulfide bonds. Structure-based bioinformatic analysis revealed independent evolutionary behaviour of both receptor domains and, together with multiple sequence alignment, identified putative binding sites for interferon-γ and receptor 1, the ligands of IFNγR2.
Organizational Affiliation:
Institute of Biotechnology CAS, BIOCEV, Prumyslova 595, 252 50 Vestec, Czech Republic.