Efficient synthesis of alpha-galactosyl oligosaccharides using a mutant Bacteroides thetaiotaomicron retaining alpha-galactosidase (BtGH97b).
Okuyama, M., Matsunaga, K., Watanabe, K.I., Yamashita, K., Tagami, T., Kikuchi, A., Ma, M., Klahan, P., Mori, H., Yao, M., Kimura, A.(2017) FEBS J 284: 766-783
- PubMed: 28103425
- DOI: https://doi.org/10.1111/febs.14018
- Primary Citation of Related Structures:
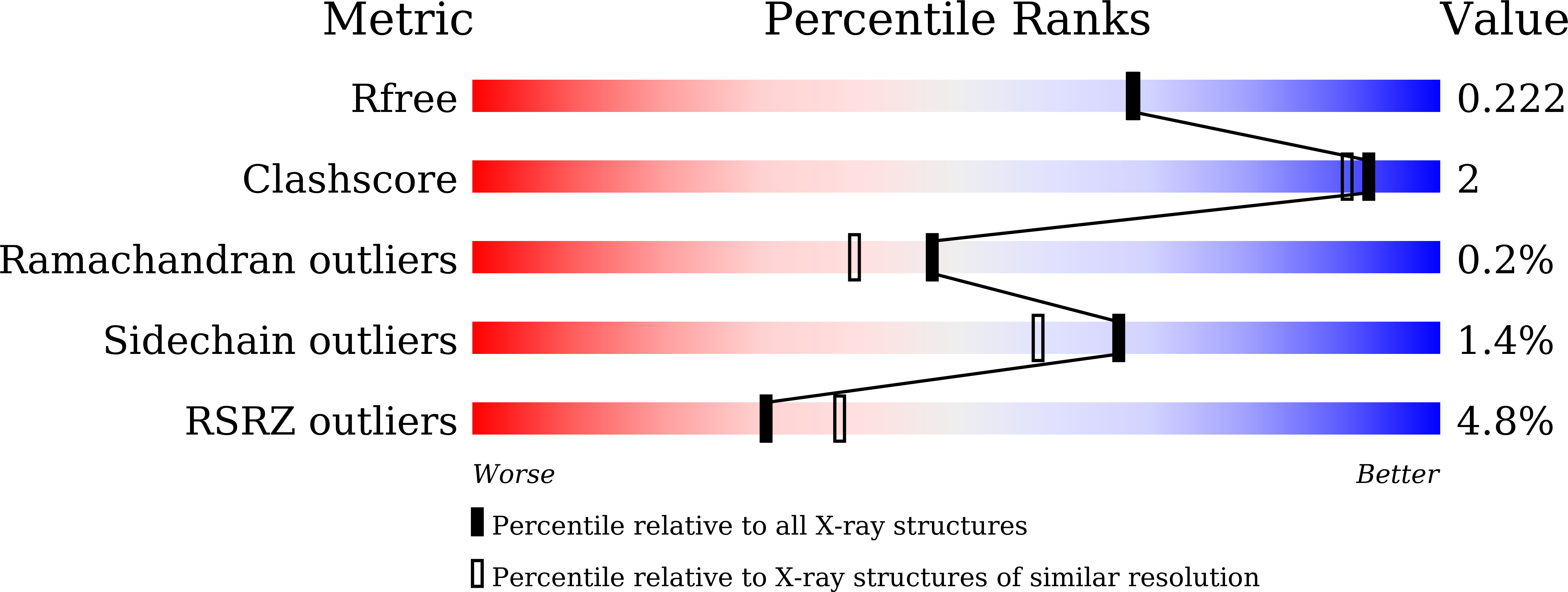
5E1Q - PubMed Abstract:
The preparation of a glycosynthase, a catalytic nucleophile mutant of a glycosidase, is a well-established strategy for the effective synthesis of glycosidic linkages. However, glycosynthases derived from α-glycosidases can give poor yields of desired products because they require generally unstable β-glycosyl fluoride donors. Here, we investigate a transglycosylation catalyzed by a catalytic nucleophile mutant derived from a glycoside hydrolase family (GH) 97 α-galactosidase, using more stable β-galactosyl azide and α-galactosyl fluoride donors. The mutant enzyme catalyzes the glycosynthase reaction using β-galactosyl azide and α-galactosyl transfer from α-galactosyl fluoride with assistance of external anions. Formate was more effective at restoring transfer activity than azide. Kinetic analysis suggests that poor transglycosylation in the presence of the azide is because of low activity of the ternary complex between enzyme, β-galactosyl azide and acceptor. A three-dimensional structure of the mutant enzyme in complex with the transglycosylation product, β-lactosyl α-d-galactoside, was solved to elucidate the ligand-binding aspects of the α-galactosidase. Subtle differences at the β→α loops 1, 2 and 3 of the catalytic TIM barrel of the α-galactosidase from those of a homologous GH97 α-glucoside hydrolase seem to be involved in substrate recognitions. In particular, the Trp residues in β→α loop 1 have separate roles. Trp312 of the α-galactosidase appears to exclude the equatorial hydroxy group at C4 of glucosides, whereas the corresponding Trp residue in the α-glucoside hydrolase makes a hydrogen bond with this hydroxy group. The mechanism of α-galactoside recognition is conserved among GH27, 31, 36 and 97 α-galactosidases. The atomic coordinates (code: 5E1Q) have been deposited in the Protein Data Bank.
Organizational Affiliation:
Laboratory of Molecular Enzymology, Research Faculty of Agriculture, Hokkaido University, Sapporo, Japan.