Unsaturated Lipid Assimilation by Mycobacteria Requires Auxiliary cis-trans Enoyl CoA Isomerase.
Srivastava, S., Chaudhary, S., Thukral, L., Shi, C., Gupta, R.D., Gupta, R., Priyadarshan, K., Vats, A., Haque, A.S., Sankaranarayanan, R., Natarajan, V.T., Sharma, R., Aldrich, C.C., Gokhale, R.S.(2015) Chem Biol 22: 1577-1587
- PubMed: 26628360
- DOI: https://doi.org/10.1016/j.chembiol.2015.10.009
- Primary Citation of Related Structures:
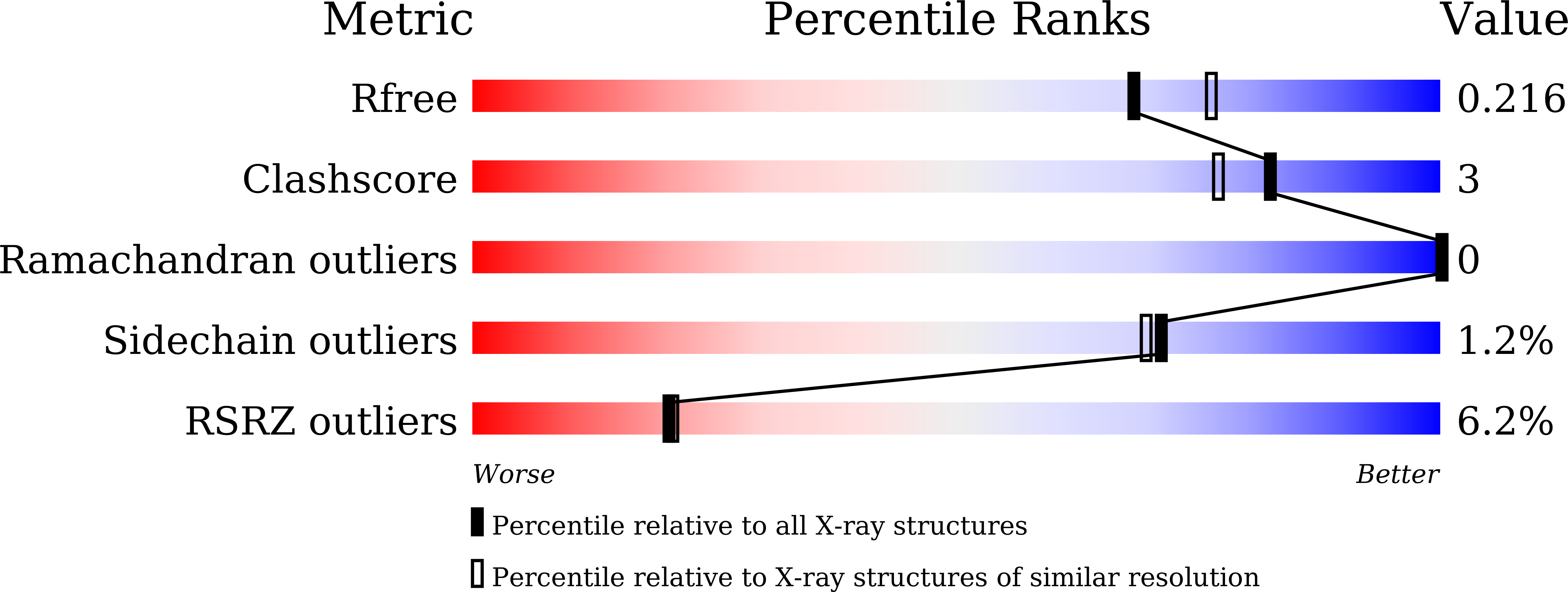
4Z0M, 5E0N - PubMed Abstract:
Mycobacterium tuberculosis (Mtb) can survive in hypoxic necrotic tissue by assimilating energy from host-derived fatty acids. While the expanded repertoire of β-oxidation auxiliary enzymes is considered crucial for Mtb adaptability, delineating their functional relevance has been challenging. Here, we show that the Mtb fatty acid degradation (FadAB) complex cannot selectively break down cis fatty acyl substrates. We demonstrate that the stereoselective binding of fatty acyl substrates in the Mtb FadB pocket is due to the steric hindrance from Phe287 residue. By developing a functional screen, we classify the family of Mtb Ech proteins as monofunctional or bifunctional enzymes, three of which complement the FadAB complex to degrade cis fatty acids. Crystal structure determination of two cis-trans enoyl coenzyme A (CoA) isomerases reveals distinct placement of active-site residue in Ech enzymes. Our studies thus reveal versatility of Mtb lipid-remodeling enzymes and identify an essential role of stand-alone cis-trans enoyl CoA isomerases in mycobacterial biology.
Organizational Affiliation:
CSIR-Institute of Genomics and Integrative Biology, Mathura Road, New Delhi 110020, India; Academy of Scientific and Innovative Research, Rafi Marg, New Delhi 110001, India.