Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus.
Lambert, C., Cadby, I.T., Till, R., Bui, N.K., Lerner, T.R., Hughes, W.S., Lee, D.J., Alderwick, L.J., Vollmer, W., Sockett, E.R., Lovering, A.L.(2015) Nat Commun 6: 8884-8884
- PubMed: 26626559
- DOI: https://doi.org/10.1038/ncomms9884
- Primary Citation of Related Structures:
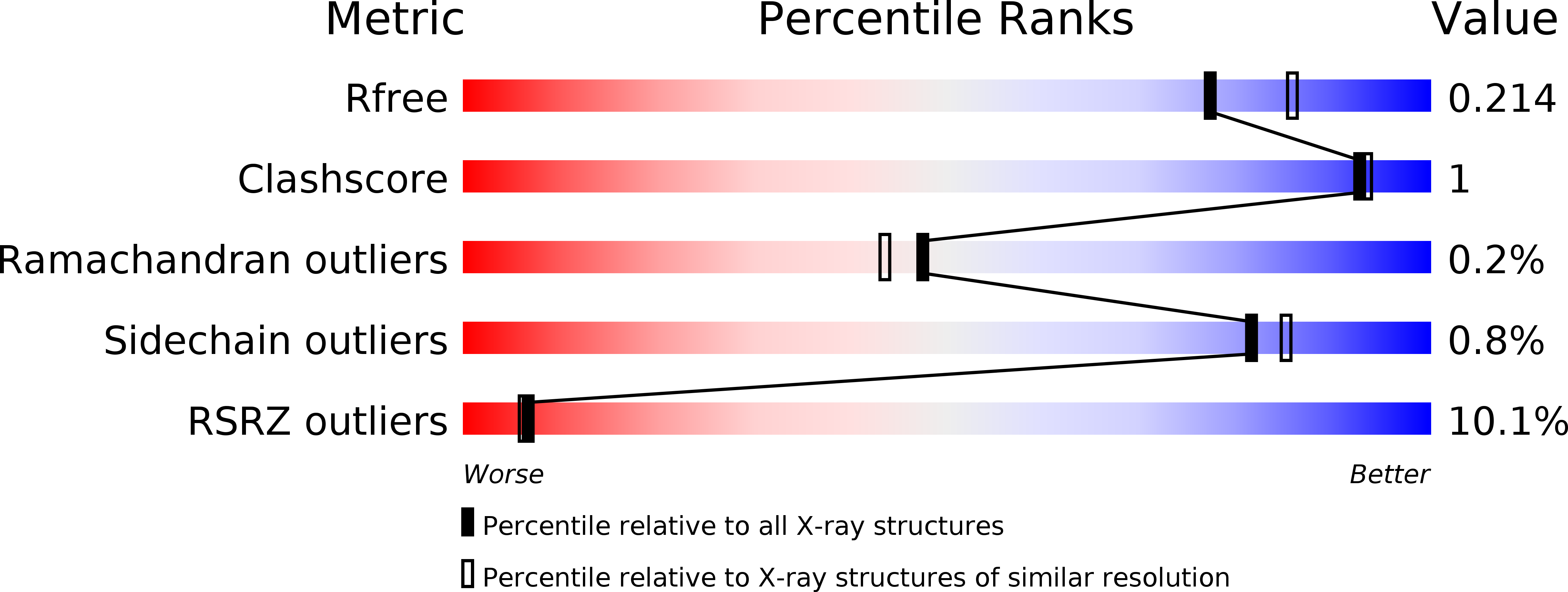
5CEA, 5CEB, 5CEC, 5CED, 5CER - PubMed Abstract:
Predatory Bdellovibrio bacteriovorus are natural antimicrobial organisms, killing other bacteria by whole-cell invasion. Self-protection against prey-metabolizing enzymes is important for the evolution of predation. Initial prey entry involves the predator's peptidoglycan DD-endopeptidases, which decrosslink cell walls and prevent wasteful entry by a second predator. Here we identify and characterize a self-protection protein from B. bacteriovorus, Bd3460, which displays an ankyrin-based fold common to intracellular pathogens of eukaryotes. Co-crystal structures reveal Bd3460 complexation of dual targets, binding a conserved epitope of each of the Bd3459 and Bd0816 endopeptidases. Complexation inhibits endopeptidase activity and cell wall decrosslinking in vitro. Self-protection is vital - ΔBd3460 Bdellovibrio deleteriously decrosslink self-peptidoglycan upon invasion, adopt a round morphology, and lose predatory capacity and cellular integrity. Our analysis provides the first mechanistic examination of self-protection in Bdellovibrio, documents protection-multiplicity for products of two different genomic loci, and reveals an important evolutionary adaptation to an invasive predatory bacterial lifestyle.
Organizational Affiliation:
Centre for Genetics and Genomics, School of Biology, Nottingham University, Medical School, Queen's Medical Centre, Nottingham NG7 2UH, UK.