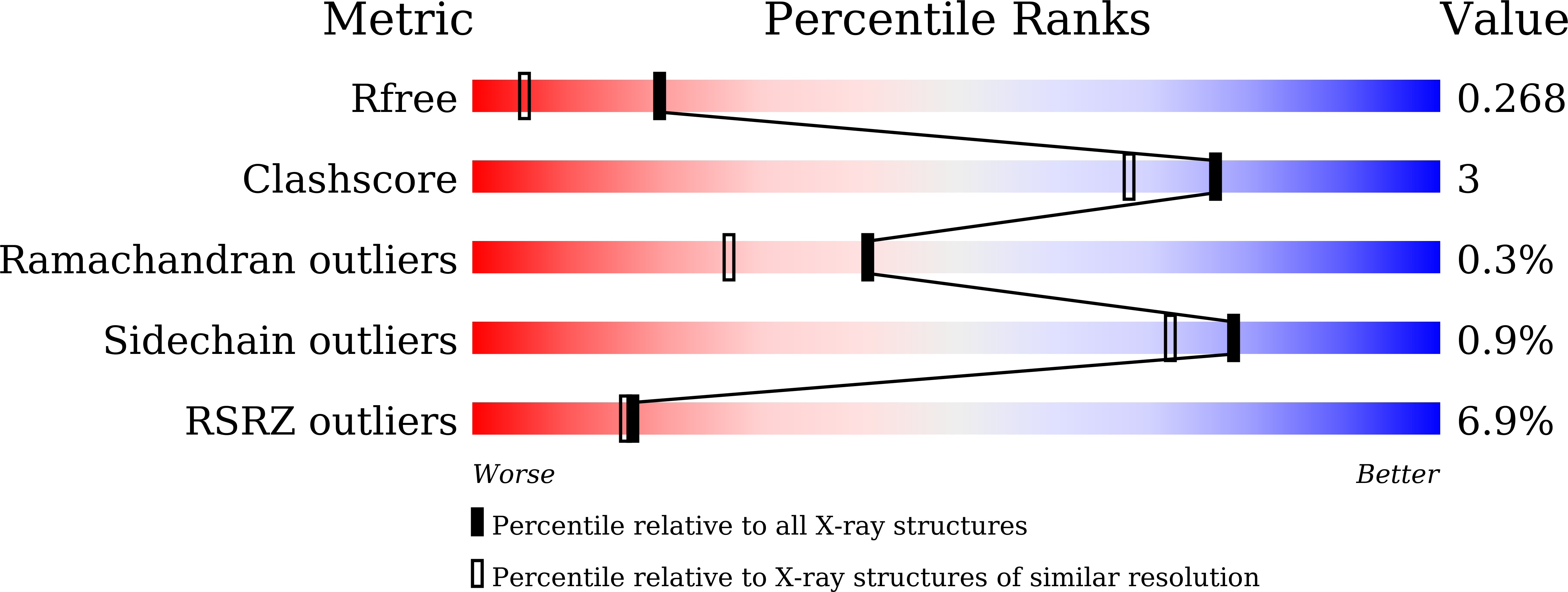
Structural and functional characterization of aspartate racemase from the acidothermophilic archaeon Picrophilus torridus
Aihara, T., Ito, T., Yamanaka, Y., Noguchi, K., Odaka, M., Sekine, M., Homma, H., Yohda, M.(2016) Extremophiles 20: 385-393
- PubMed: 27094682
- DOI: https://doi.org/10.1007/s00792-016-0829-7
- Primary Citation of Related Structures:
5B19 - PubMed Abstract:
Functional and structural characterizations of pyridoxal 5'-phosphate-independent aspartate racemase of the acidothermophilic archaeon Picrophilus torridus were performed. Picrophilus aspartate racemase exhibited high substrate specificity to aspartic acid. The optimal reaction temperature was 60 °C, which is almost the same as the optimal growth temperature. Reflecting the low pH in the cytosol, the optimal reaction pH of Picrophilus aspartate racemase was approximately 5.5. However, the activity at the putative cytosolic pH of 4.6 was approximately 6 times lower than that at the optimal pH of 5.5. The crystal structure of Picrophilus aspartate racemase was almost the same as that of other pyridoxal 5'-phosphate -independent aspartate racemases. In two molecules of the dimer, one molecule contained a tartaric acid molecule in the catalytic site; the structure of the other molecule was relatively flexible. Finally, we examined the intracellular existence of D-amino acids. Unexpectedly, the proportion of D-aspartate to total aspartate was not very high. In contrast, both D-proline and D-alanine were observed. Because Picrophilus aspartate racemase is highly specific to aspartate, other amino acid racemases might exist in Picrophilus torridus.
Organizational Affiliation:
Department of Biotechnology and Life Science, Graduate School of Technology, Tokyo University of Agriculture and Technology, 2-24-16 Naka-cho, Koganei, Tokyo, 184-8588, Japan.