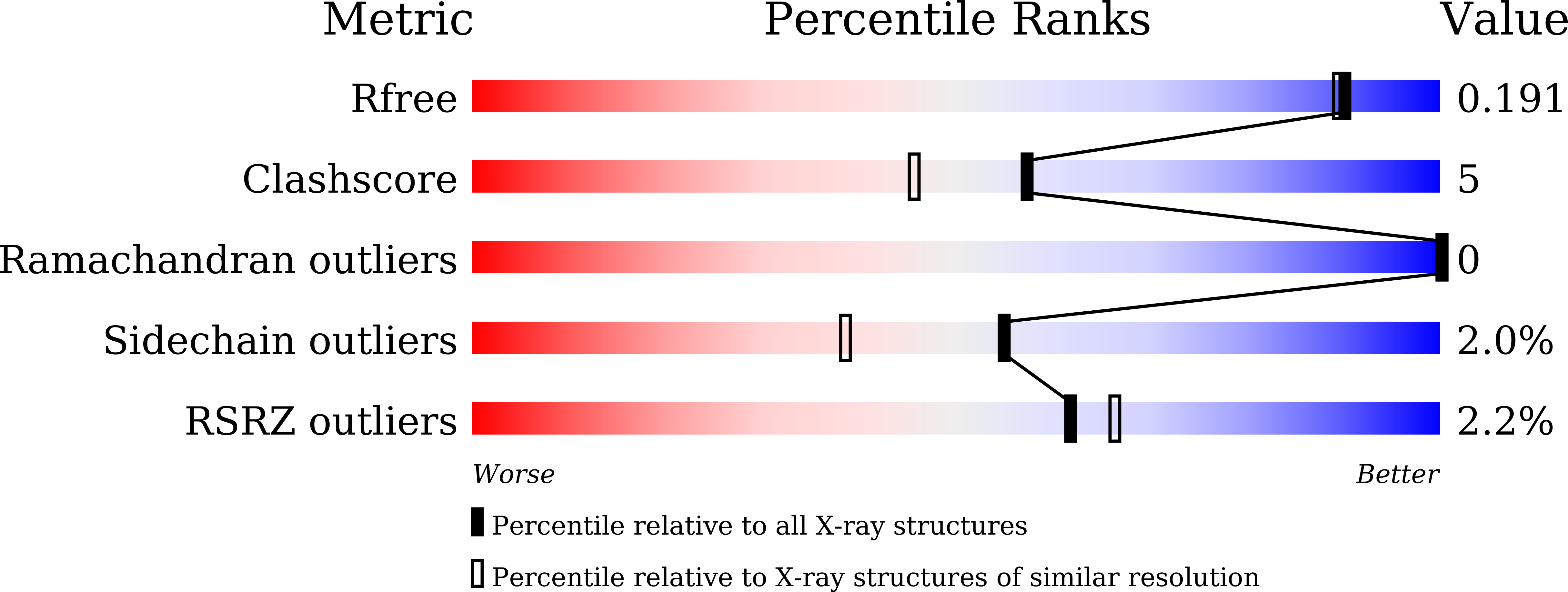
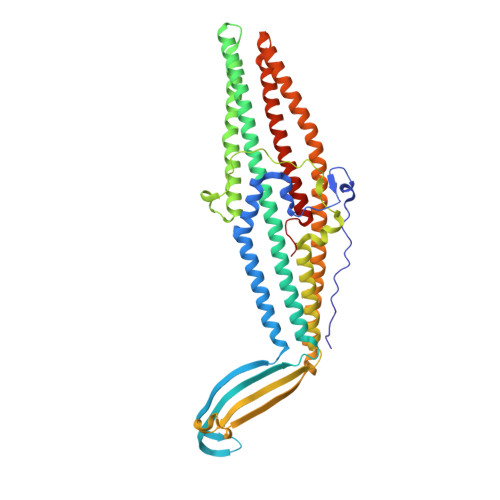
Crystal structures of OprN and OprJ, outer membrane factors of multidrug tripartite efflux pumps of Pseudomonas aeruginosa.
Yonehara, R., Yamashita, E., Nakagawa, A.(2016) Proteins 84: 759-769
- PubMed: 26914226
- DOI: https://doi.org/10.1002/prot.25022
- Primary Citation of Related Structures:
5AZO, 5AZP, 5AZS - PubMed Abstract:
The genome of Pseudomonas aeruginosa encodes tripartite efflux pumps that extrude functionally and structurally dissimilar antibiotics from the bacterial cell. MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM are the main tripartite efflux pumps responsible for multidrug resistance in P. aeruginosa. The outer membrane factors OprN, OprJ, and OprM are essential components of functional tripartite efflux pumps. To elucidate the structural basis of multidrug resistance, we determined the crystal structures of OprN and OprJ. These structures revealed several features, including tri-acylation of the N-terminal cysteine, a small pore in the β-barrel domain, and a tightly sealed gate in the α-barrel domain. Despite the overall similarity of OprN, OprJ, and OprM, a comparison of their structures and electrostatic distributions revealed subtle differences at the periplasmic end of the α-barrel domain. These results suggested that the overall structures of these outer membrane factors are specifically optimized for particular tripartite efflux pumps. Proteins 2016; 84:759-769. © 2016 Wiley Periodicals, Inc.
Organizational Affiliation:
Institute for Protein Research, Osaka University, Suita, Japan.