The Use of Noncrystallographic Symmetry Averaging to Solve Structures from Data Affected by Perfect Hemihedral Twinning
Sabin, C., Plevka, P.(2016) Acta Crystallogr Sect F Struct Biol Cryst Commun 72: 188
- PubMed: 26919522
- DOI: https://doi.org/10.1107/S2053230X16000923
- Primary Citation of Related Structures:
5AOO - PubMed Abstract:
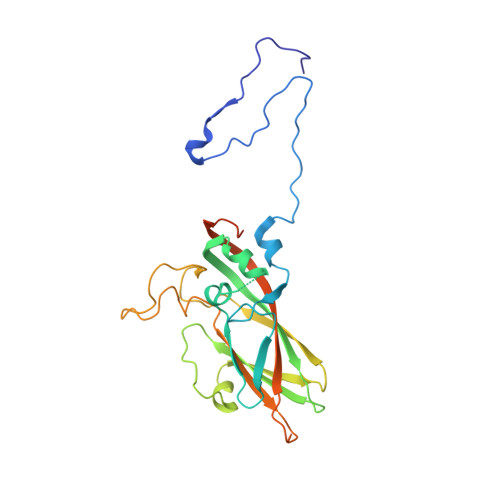
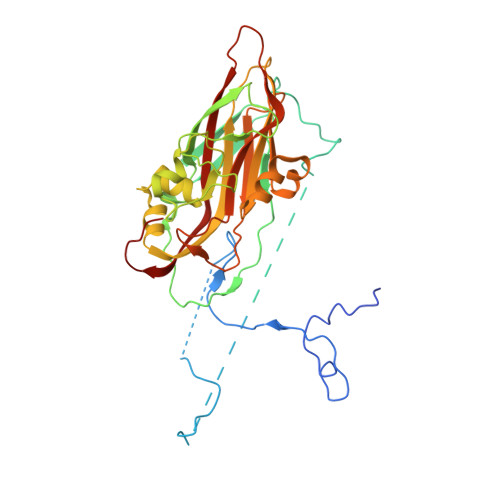
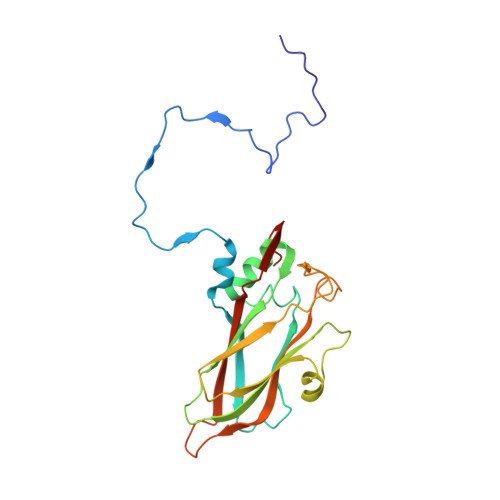
Hemihedral twinning is a crystal-growth anomaly in which a specimen is composed of two crystal domains that coincide with each other in three dimensions. However, the orientations of the crystal lattices in the two domains differ in a specific way. In diffraction data collected from hemihedrally twinned crystals, each observed intensity contains contributions from both of the domains. With perfect hemihedral twinning, the two domains have the same volumes and the observed intensities do not contain sufficient information to detwin the data. Here, the use of molecular replacement and of noncrystallographic symmetry (NCS) averaging to detwin a 2.1 Å resolution data set for Aichi virus 1 affected by perfect hemihedral twinning is described. The NCS averaging enabled the correction of errors in the detwinning introduced by the differences between the molecular-replacement model and the crystallized structure. The procedure permitted the structure to be determined from a molecular-replacement model that had 16% sequence identity and a 1.6 Å r.m.s.d. for C(α) atoms in comparison to the crystallized structure. The same approach could be used to solve other data sets affected by perfect hemihedral twinning from crystals with NCS.
Organizational Affiliation:
Central European Institute of Technology - Masaryk University, Kamenice 653/25, 625 00 Brno, Czech Republic.