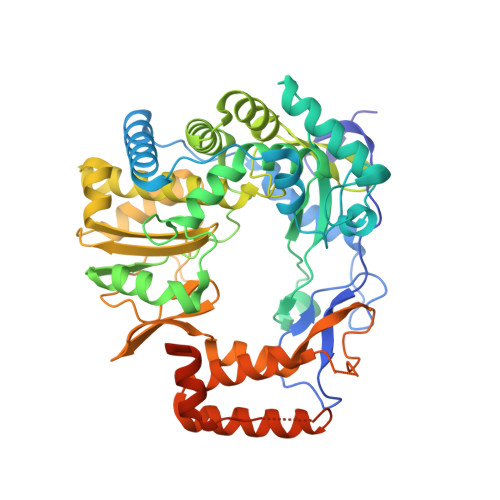
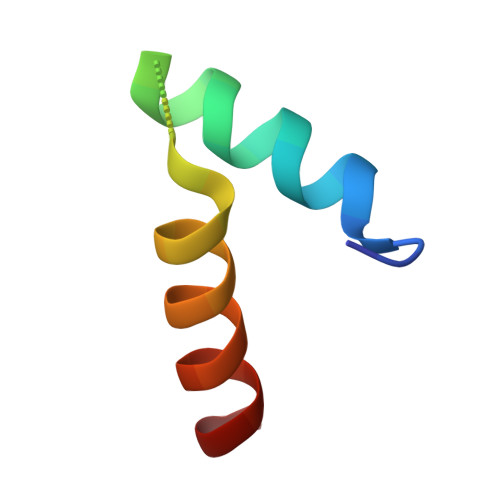
Insight Into the Interaction Between RNA Polymerase and VPg for Murine Norovirus Replication.
Lee, J.H., Park, B.S., Han, K.R., Biering, S.B., Kim, S.J., Choi, J., Seok, J.H., Alam, I., Chung, M.S., Kim, H.M., Hwang, S., Kim, K.H.(2018) Front Microbiol 9: 1466-1466
- PubMed: 30038601
- DOI: https://doi.org/10.3389/fmicb.2018.01466
- Primary Citation of Related Structures:
5Y3D - PubMed Abstract:
Norovirus (NoV) is a leading cause of epidemic acute non-bacterial gastroenteritis, and replicates through virion protein genome-linked (VPg)-primed or de novo RNA synthesis by RNA-dependent RNA polymerase (RdRp). VPg is a multifunctional protein that plays crucial roles in viral protein translation and genome replication. However, the interaction between the RdRp and this multifunctional VPg in NoV replication has been unknown. In this study, VPg derived from murine NoV (MNV) was found to mediate the formation of higher-order multimers or tubular fibrils of MNV RdRp, which led to significantly enhanced polymerase activity in vitro . The replication of MNV mutants containing a VPg-binding defective RdRp, based on the crystal structure of an RdRp-VPg(1-73) complex, was completely blocked in a cell culture system. Our data suggest that the interaction between RdRp and VPg plays a crucial role in the multimerization-mediated RdRp activity in vivo and consequently in MNV replication, which can provide a new target of therapeutic intervention for NoV outbreaks.
Organizational Affiliation:
Department of Biotechnology & Bioinformatics, Korea University, Sejong, South Korea.