Structural Insight into the Activation of PknI Kinase from M. tuberculosis via Dimerization of the Extracellular Sensor Domain.
Yan, Q., Jiang, D., Qian, L., Zhang, Q., Zhang, W., Zhou, W., Mi, K., Guddat, L., Yang, H., Rao, Z.(2017) Structure 25: 1286-1294.e4
- PubMed: 28712808
- DOI: https://doi.org/10.1016/j.str.2017.06.010
- Primary Citation of Related Structures:
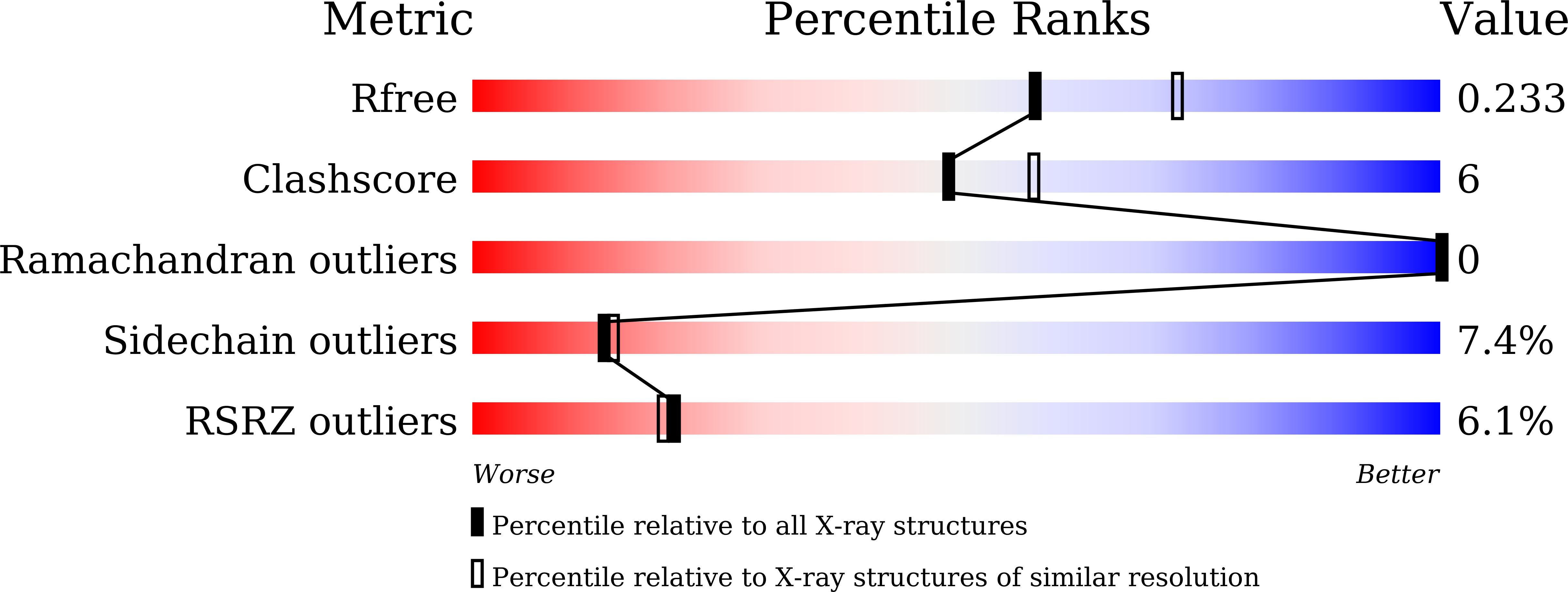
5XKA, 5XLL, 5XLM - PubMed Abstract:
Protein kinases play central roles in the survival of Mycobacterium tuberculosis within host. Here we report the individual high-resolution crystal structures of the sensor domain (in both monomer and dimer forms) and the kinase domain of PknI, a transmembrane protein member of the serine/threonine protein kinases (STPKs) family. PknI is the first STPK identified whose sensor domain exists in a monomer-dimer equilibrium. Inspection of the two structures of the sensor domain (PknI_SD) revealed conformational changes upon dimerization, with an arm region of critical importance for dimer formation identified. Rapamycin-induced dimerization of unphosphorylated fusions of PknI juxtamembrane and the kinase domain, intended to mimic the dimerization effect presumably imposed by PknI_SD, was observed to be able to activate auto-phosphorylation activity of the kinase domain. In vivo experiments using an M. bovis model suggested PknI functions as a dimer in the regulation of M. tuberculosis growth.
Organizational Affiliation:
College of Life Sciences, Nankai University, Tianjin 300071, China.