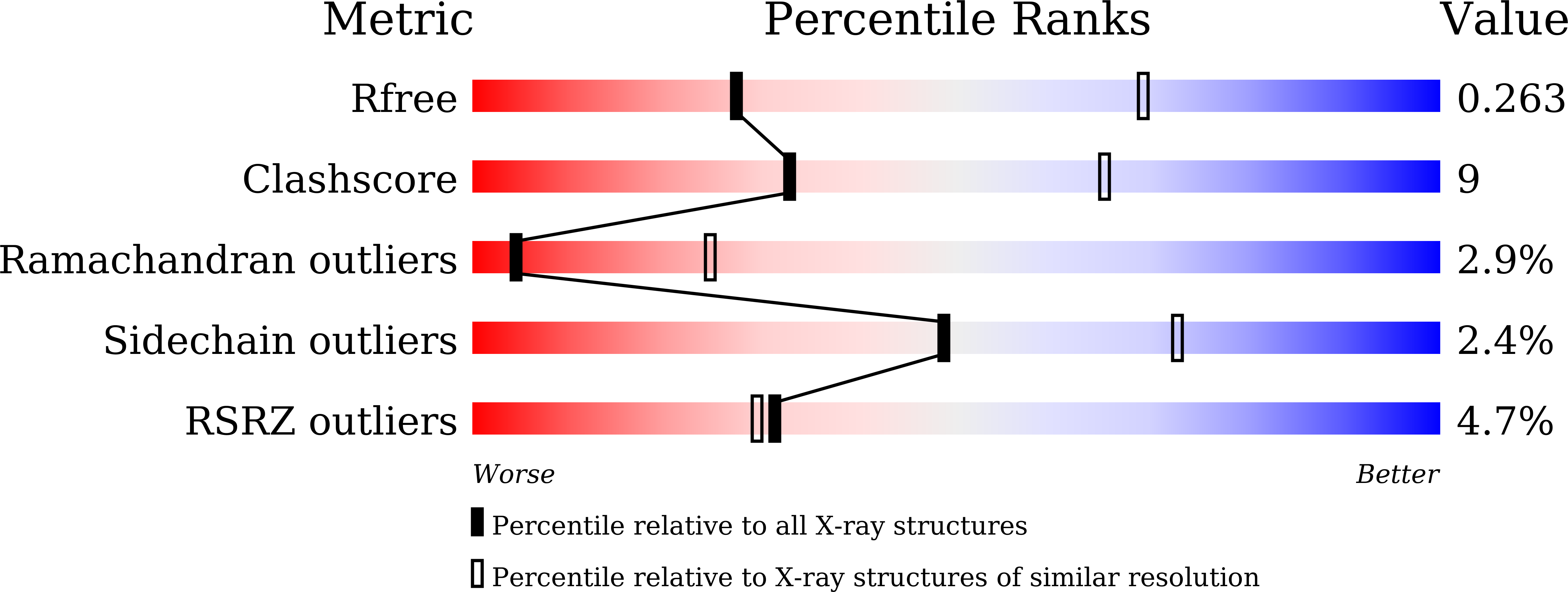
Mechanism of formation of a toroid around DNA by the mismatch sensor protein.
Nirwal, S., Kulkarni, D.S., Sharma, A., Rao, D.N., Nair, D.T.(2018) Nucleic Acids Res 46: 256-266
- PubMed: 29182773
- DOI: https://doi.org/10.1093/nar/gkx1149
- Primary Citation of Related Structures:
5X9W, 5YK4 - PubMed Abstract:
The DNA mismatch repair (MMR) pathway removes errors that appear during genome replication. MutS is the primary mismatch sensor and forms an asymmetric dimer that encircles DNA to bend it to scan for mismatches. The mechanism utilized to load DNA into the central tunnel was unknown and the origin of the force required to bend DNA was unclear. We show that, in absence of DNA, MutS forms a symmetric dimer wherein a gap exists between the monomers through which DNA can enter the central tunnel. The comparison with structures of MutS-DNA complexes suggests that the mismatch scanning monomer (Bm) will move by nearly 50 Å to associate with the other monomer (Am). Consequently, the N-terminal domains of both monomers will press onto DNA to bend it. The proposed mechanism of toroid formation evinces that the force required to bend DNA arises primarily due to the movement of Bm and hence, the MutS dimer acts like a pair of pliers to bend DNA. We also shed light on the allosteric mechanism that influences the expulsion of adenosine triphosphate from Am on DNA binding. Overall, this study provides mechanistic insight regarding the primary event in MMR i.e. the assembly of the MutS-DNA complex.
Organizational Affiliation:
Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Faridabad-Gurgaon Expressway, Faridabad, 121001, Haryana, India.