Neutron and Atomic Resolution X-ray Structures of a Lytic Polysaccharide Monooxygenase Reveal Copper-Mediated Dioxygen Binding and Evidence for N-Terminal Deprotonation.
Bacik, J.P., Mekasha, S., Forsberg, Z., Kovalevsky, A.Y., Vaaje-Kolstad, G., Eijsink, V.G.H., Nix, J.C., Coates, L., Cuneo, M.J., Unkefer, C.J., Chen, J.C.(2017) Biochemistry 56: 2529-2532
- PubMed: 28481095
- DOI: https://doi.org/10.1021/acs.biochem.7b00019
- Primary Citation of Related Structures:
5VG0, 5VG1 - PubMed Abstract:
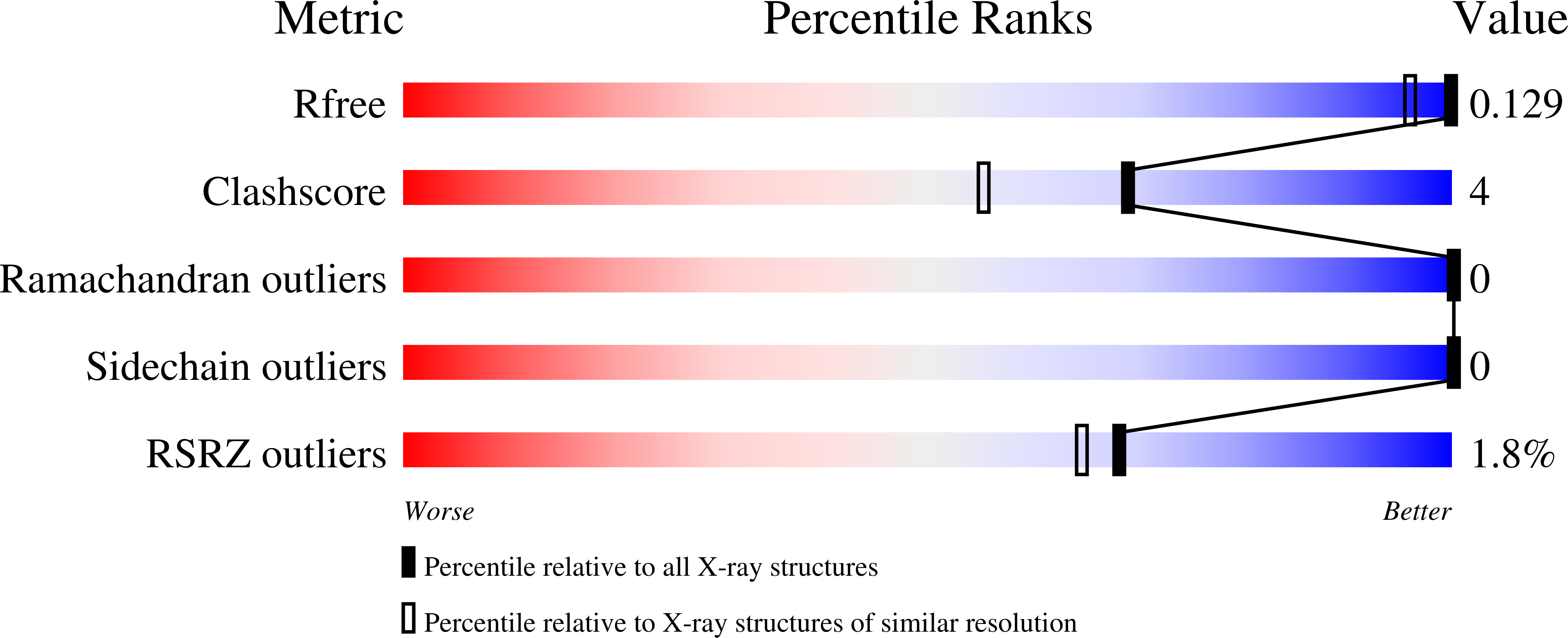
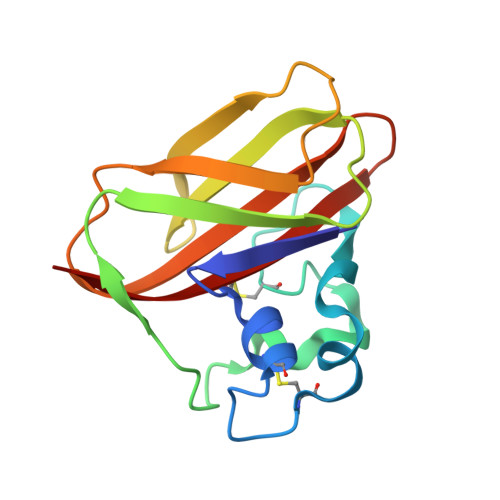
A 1.1 Å resolution, room-temperature X-ray structure and a 2.1 Å resolution neutron structure of a chitin-degrading lytic polysaccharide monooxygenase domain from the bacterium Jonesia denitrificans (JdLPMO10A) show a putative dioxygen species equatorially bound to the active site copper. Both structures show an elongated density for the dioxygen, most consistent with a Cu(II)-bound peroxide. The coordination environment is consistent with Cu(II). In the neutron and X-ray structures, difference maps reveal the N-terminal amino group, involved in copper coordination, is present as a mixed ND 2 and ND - , suggesting a role for the copper ion in shifting the pK a of the amino terminus.
Organizational Affiliation:
Protein Crystallography Station, Bioscience Division, Los Alamos National Laboratory , Los Alamos, New Mexico 87545, United States.