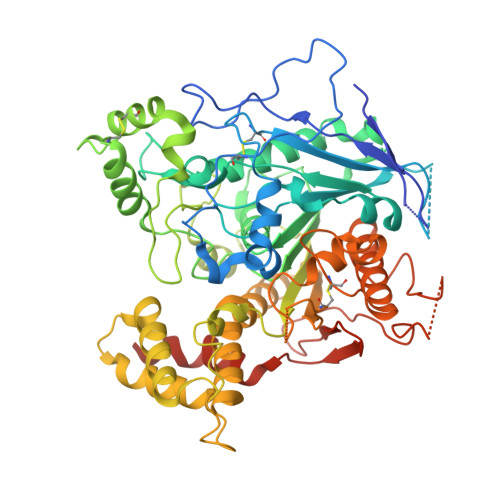
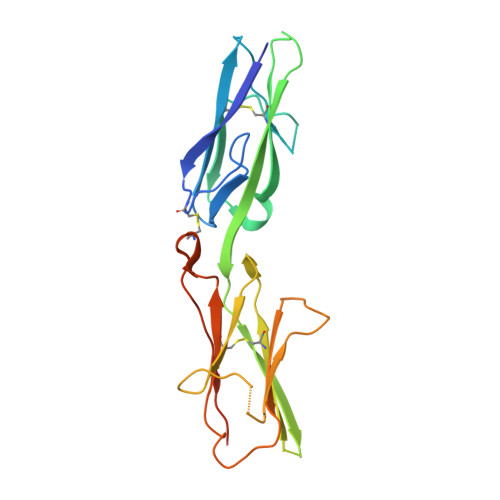
Molecular Mechanism of MDGA1: Regulation of Neuroligin 2:Neurexin Trans-synaptic Bridges.
Gangwar, S.P., Zhong, X., Seshadrinathan, S., Chen, H., Machius, M., Rudenko, G.(2017) Neuron 94: 1132-1141.e4
- PubMed: 28641112
- DOI: https://doi.org/10.1016/j.neuron.2017.06.009
- Primary Citation of Related Structures:
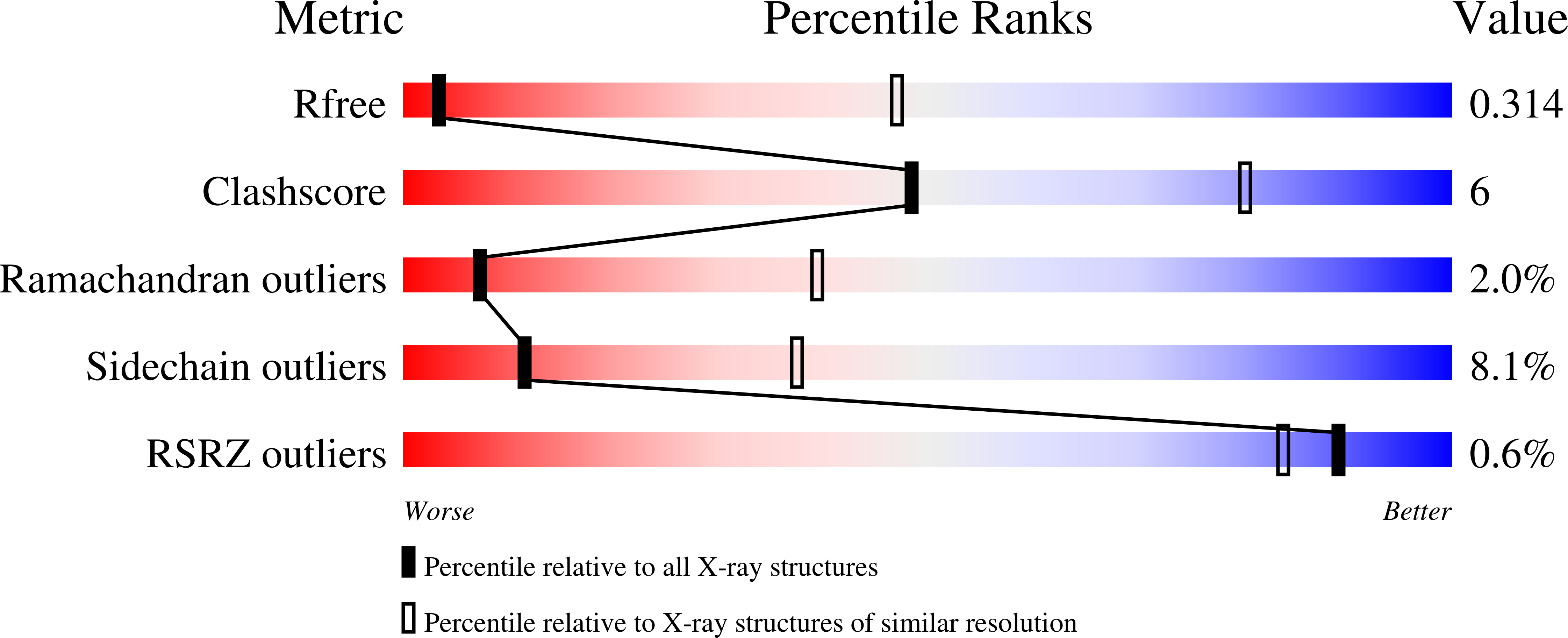
5V5V, 5V5W - PubMed Abstract:
Neuroligins and neurexins promote synapse development and validation by forming trans-synaptic bridges spanning the synaptic cleft. Select pairs promote excitatory and inhibitory synapses, with neuroligin 2 (NLGN2) limited to inhibitory synapses and neuroligin 1 (NLGN1) dominating at excitatory synapses. The cell-surface molecules, MAM domain-containing glycosylphosphatidylinositol anchor 1 (MDGA1) and 2 (MDGA2), regulate trans-synaptic adhesion between neurexins and neuroligins, impacting NLGN2 and NLGN1, respectively. We have determined the molecular mechanism of MDGA action. MDGA1 Ig1-Ig2 is sufficient to bind NLGN2 with nanomolar affinity; its crystal structure reveals an unusual locked rod-shaped array. In the crystal structure of the complex, two MDGA1 Ig1-Ig2 molecules each span the entire NLGN2 dimer. Site-directed mutagenesis confirms the observed interaction interface. Strikingly, Ig1 from MDGA1 binds to the same region on NLGN2 as neurexins do. Thus, MDGAs regulate the formation of neuroligin-neurexin trans-synaptic bridges by sterically blocking access of neurexins to neuroligins.
Organizational Affiliation:
Department of Pharmacology and Toxicology, University of Texas Medical Branch, Galveston, TX 77555, USA; Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, Galveston, TX 77555, USA.