Denisovan, modern human and mouse TNFAIP3 alleles tune A20 phosphorylation and immunity.
Zammit, N.W., Siggs, O.M., Gray, P.E., Horikawa, K., Langley, D.B., Walters, S.N., Daley, S.R., Loetsch, C., Warren, J., Yap, J.Y., Cultrone, D., Russell, A., Malle, E.K., Villanueva, J.E., Cowley, M.J., Gayevskiy, V., Dinger, M.E., Brink, R., Zahra, D., Chaudhri, G., Karupiah, G., Whittle, B., Roots, C., Bertram, E., Yamada, M., Jeelall, Y., Enders, A., Clifton, B.E., Mabbitt, P.D., Jackson, C.J., Watson, S.R., Jenne, C.N., Lanier, L.L., Wiltshire, T., Spitzer, M.H., Nolan, G.P., Schmitz, F., Aderem, A., Porebski, B.T., Buckle, A.M., Abbott, D.W., Ziegler, J.B., Craig, M.E., Benitez-Aguirre, P., Teo, J., Tangye, S.G., King, C., Wong, M., Cox, M.P., Phung, W., Tang, J., Sandoval, W., Wertz, I.E., Christ, D., Goodnow, C.C., Grey, S.T.(2019) Nat Immunol 20: 1299-1310
- PubMed: 31534238
- DOI: https://doi.org/10.1038/s41590-019-0492-0
- Primary Citation of Related Structures:
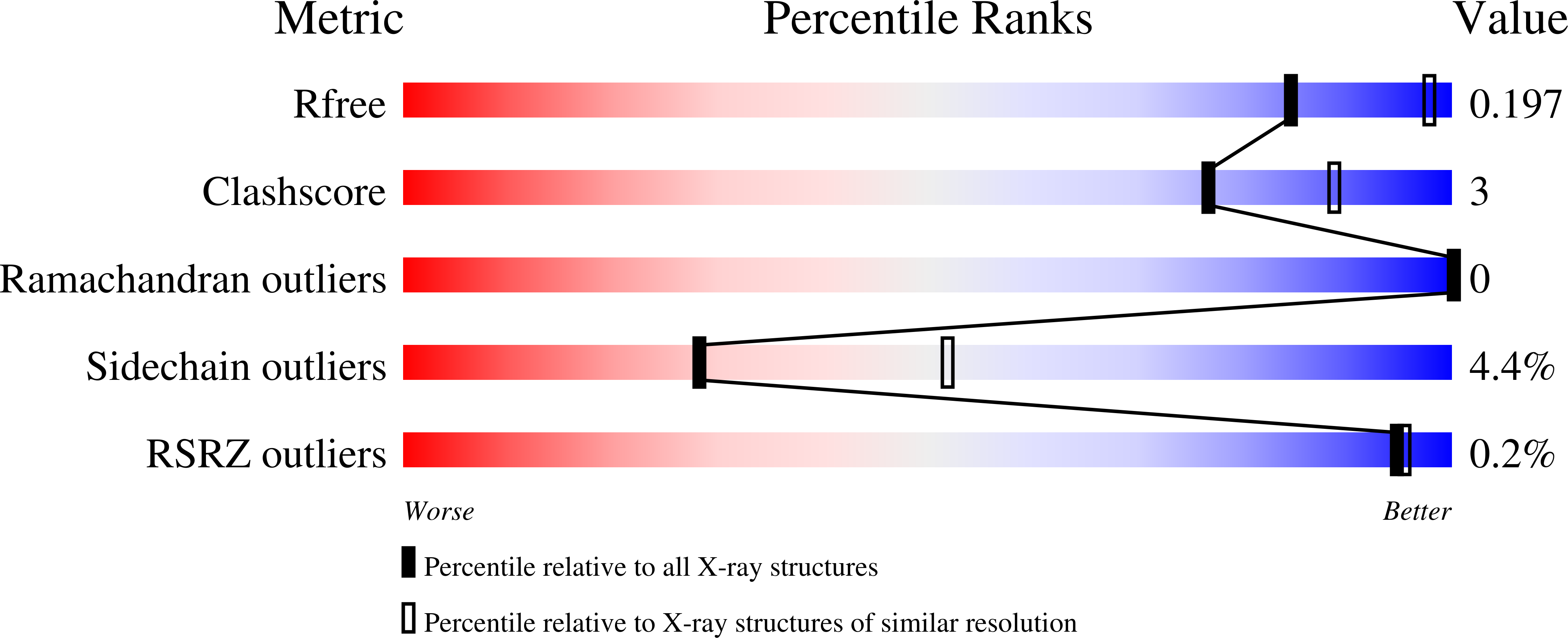
5V3B, 5V3P - PubMed Abstract:
Resisting and tolerating microbes are alternative strategies to survive infection, but little is known about the evolutionary mechanisms controlling this balance. Here genomic analyses of anatomically modern humans, extinct Denisovan hominins and mice revealed a TNFAIP3 allelic series with alterations in the encoded immune response inhibitor A20. Each TNFAIP3 allele encoded substitutions at non-catalytic residues of the ubiquitin protease OTU domain that diminished IκB kinase-dependent phosphorylation and activation of A20. Two TNFAIP3 alleles encoding A20 proteins with partial phosphorylation deficits seemed to be beneficial by increasing immunity without causing spontaneous inflammatory disease: A20 T108A;I207L, originating in Denisovans and introgressed in modern humans throughout Oceania, and A20 I325N, from an N-ethyl-N-nitrosourea (ENU)-mutagenized mouse strain. By contrast, a rare human TNFAIP3 allele encoding an A20 protein with 95% loss of phosphorylation, C243Y, caused spontaneous inflammatory disease in humans and mice. Analysis of the partial-phosphorylation A20 I325N allele in mice revealed diminished tolerance of bacterial lipopolysaccharide and poxvirus inoculation as tradeoffs for enhanced immunity.
Organizational Affiliation:
Garvan Institute of Medical Research, Darlinghurst, New South Wales, Australia.