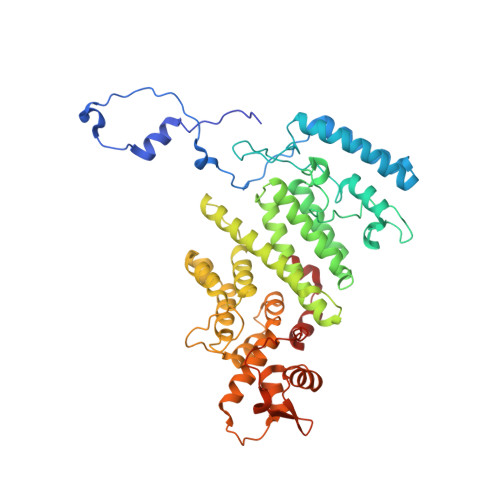
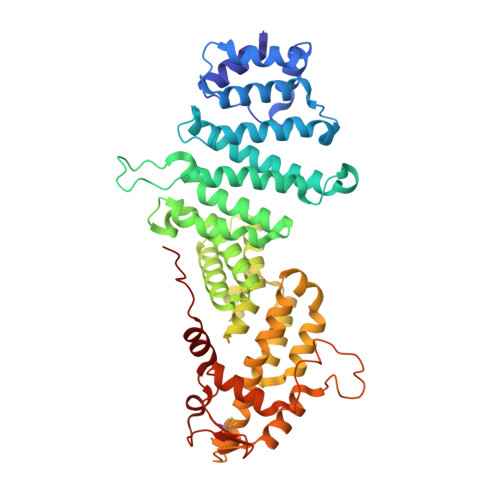
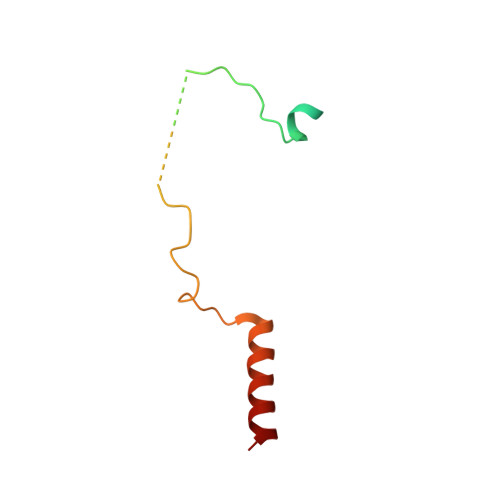
Structure of the Sac3 RNA-binding M-region in the Saccharomyces cerevisiae TREX-2 complex.
Gordon, J.M.B., Aibara, S., Stewart, M.(2017) Nucleic Acids Res 45: 5577-5585
- PubMed: 28334829
- DOI: https://doi.org/10.1093/nar/gkx158
- Primary Citation of Related Structures:
5UBP - PubMed Abstract:
Transcription-export complex 2 (TREX-2, or THSC) facilitates localization of actively transcribing genes such as GAL1 to the nuclear periphery, contributes to the generation of export-competent mRNPs and influences gene expression through interactions with Mediator. TREX-2 is based on a Sac3 scaffold to which Thp1, Sem1, Cdc31 and Sus1 bind and consists of three modules: the N-region (Sac3∼1-100), which binds mRNA export factor Mex67:Mtr2; the M-region, in which Thp1 and Sem1 bind to Sac3∼100-550; and the CID region in which Cdc31 and two Sus1 chains bind to Sac3∼720-805. Although the M-region of Sac3 was originally thought to encompass residues ∼250-550, we report here the 2.3Å resolution crystal structure of a complex containing Sac3 residues 60-550 that indicates that the TPR-like repeats of the M-region extend to residue 137 and that residues 90-125 form a novel loop that links Sac3 to Thp1. These new structural elements are important for growth and mRNA export in vivo. Although deleting Sac3 residues 1-90 produced a wild-type phenotype, deletion of the loop as well generated growth defects at 37°C, whereas the deletion of residues 1-250 impaired mRNA export and also generated longer lag times when glucose or raffinose was replaced by galactose as the carbon source.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge CB2 0QH, UK.