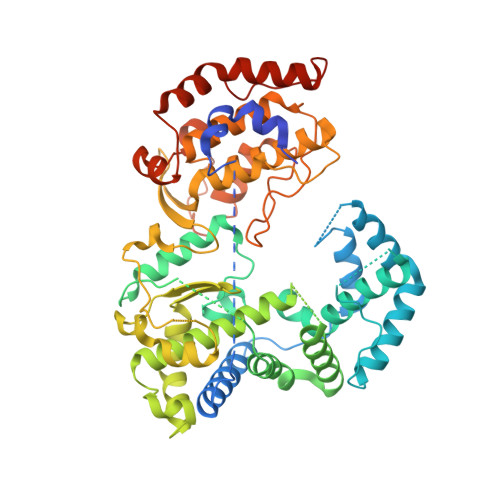
Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase.
Godoy, A.S., Lima, G.M., Oliveira, K.I., Torres, N.U., Maluf, F.V., Guido, R.V., Oliva, G.(2017) Nat Commun 8: 14764-14764
- PubMed: 28345596
- DOI: https://doi.org/10.1038/ncomms14764
- Primary Citation of Related Structures:
5U04 - PubMed Abstract:
The current Zika virus (ZIKV) outbreak became a global health threat of complex epidemiology and devastating neurological impacts, therefore requiring urgent efforts towards the development of novel efficacious and safe antiviral drugs. Due to its central role in RNA viral replication, the non-structural protein 5 (NS5) RNA-dependent RNA-polymerase (RdRp) is a prime target for drug discovery. Here we describe the crystal structure of the recombinant ZIKV NS5 RdRp domain at 1.9 Å resolution as a platform for structure-based drug design strategy. The overall structure is similar to other flaviviral homologues. However, the priming loop target site, which is suitable for non-nucleoside polymerase inhibitor design, shows significant differences in comparison with the dengue virus structures, including a tighter pocket and a modified local charge distribution.
Organizational Affiliation:
Institute of Physics of São Carlos, University of São Paulo, Av. Joao Dagnone, 1100-Jardim Santa Angelina, São Carlos 13563-120, Brazil.