Crystal structure of Pisum arvense seed lectin (PAL) and characterization of its interaction with carbohydrates by molecular docking and dynamics.
Pinto-Junior, V.R., Santiago, M.Q., Nobre, C.B., Osterne, V.J.S., Leal, R.B., Cajazeiras, J.B., Lossio, C.F., Rocha, B.A.M., Martins, M.G.Q., Nobre, C.A.S., Silva, M.T.L., Nascimento, K.S., Cavada, B.S.(2017) Arch Biochem Biophys 630: 27-37
- PubMed: 28754321
- DOI: https://doi.org/10.1016/j.abb.2017.07.013
- Primary Citation of Related Structures:
5T7P - PubMed Abstract:
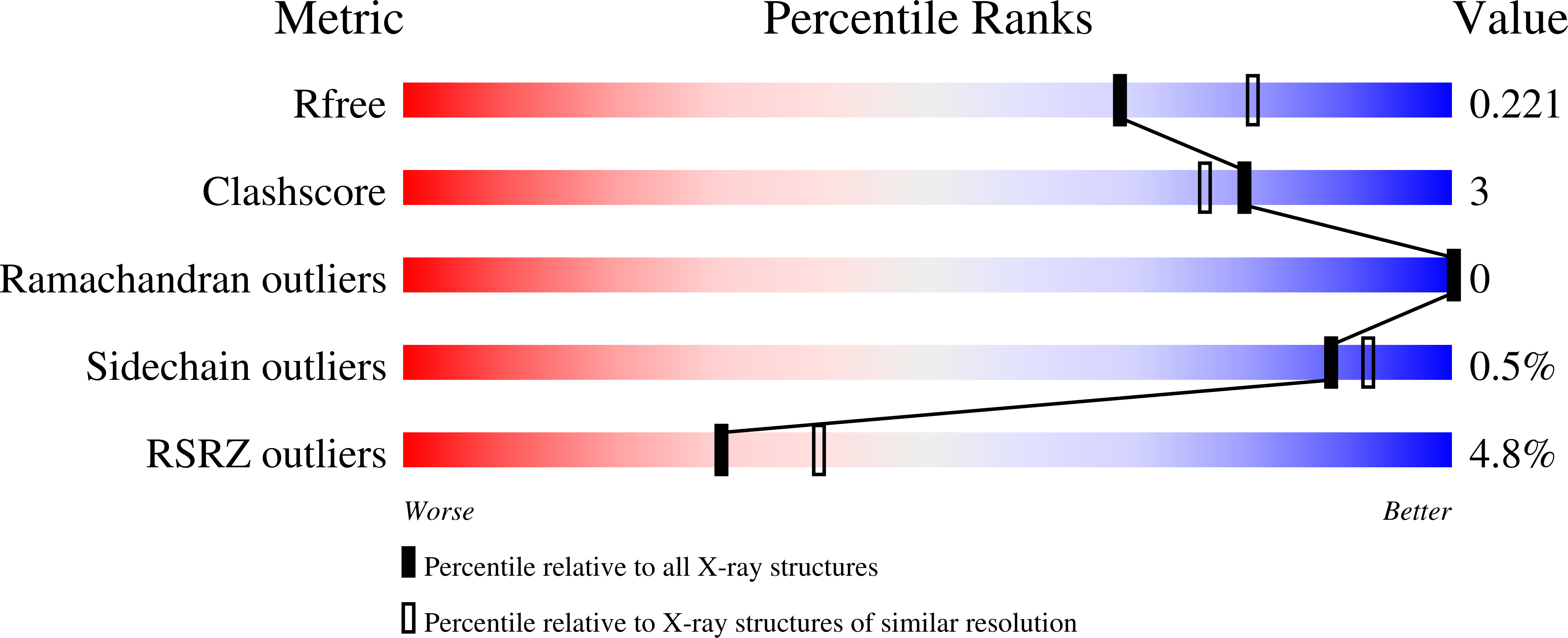
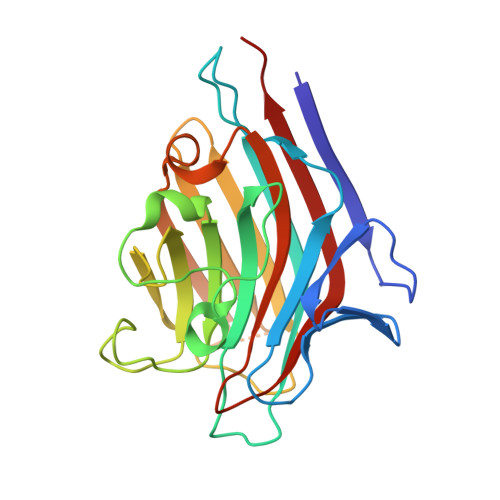
The Pisum arvense lectin (PAL), a legume protein belonging to the Vicieae tribe, is capable of specific recognition of mannose, glucose and its derivatives without altering its structure. In this work, the three-dimensional structure of PAL was determined by X-ray crystallography and studied in detail by a combination of molecular docking and molecular dynamics (MD). Crystals belonging to monoclinic space group P2 1 were grown by the vapor diffusion method at 293 K. The structure was solved at 2.16 Å and was similar to that of other Vicieae lectins. The structure presented R factor and R free of 17.04% and 22.08%, respectively, with all acceptable geometric parameters. Molecular docking was performed to analyze interactions of the lectin with monosaccharides, disaccharides and high-mannose N-glycans. PAL demonstrated different affinities on carbohydrates, depending on bond orientation and glycosidic linkage present in ligands. Furthermore, the lectin interacted with representative N-glycans in a manner consistent with the biological effects described for Vicieae lectins. Carbohydrate-recognition domain (CRD) in-depth analysis was performed by MD, describing the behavior of CRD residues in complex with ligand, stability, flexibility of the protein over time, CRD volume and topology. This is a first report of its kind for a lectin of the Vicieae tribe.
Organizational Affiliation:
Universidade Federal do Ceara (UFC), Fortaleza, Ceara, Brazil.