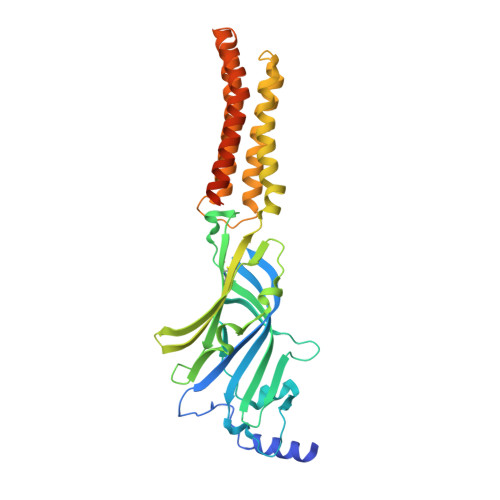
Structural basis for GABAA receptor potentiation by neurosteroids.
Miller, P.S., Scott, S., Masiulis, S., De Colibus, L., Pardon, E., Steyaert, J., Aricescu, A.R.(2017) Nat Struct Mol Biol 24: 986-992
- PubMed: 28991263
- DOI: https://doi.org/10.1038/nsmb.3484
- Primary Citation of Related Structures:
5O8F, 5OJM - PubMed Abstract:
Type A γ-aminobutyric acid receptors (GABA A Rs) are the principal mediators of inhibitory neurotransmission in the human brain. Endogenous neurosteroids interact with GABA A Rs to regulate acute and chronic anxiety and are potent sedative, analgesic, anticonvulsant and anesthetic agents. Their mode of binding and mechanism of receptor potentiation, however, remain unknown. Here we report crystal structures of a chimeric GABA A R construct in apo and pregnanolone-bound states. The neurosteroid-binding site is mechanically coupled to the helices lining the ion channel pore and modulates the desensitization-gate conformation. We demonstrate that the equivalent site is responsible for physiological, heteromeric GABA A R potentiation and explain the contrasting modulatory properties of 3a versus 3b neurosteroid epimers. These results illustrate how peripheral lipid ligands can regulate the desensitization gate of GABA A Rs, a process of broad relevance to pentameric ligand-gated ion channels.
Organizational Affiliation:
Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK.