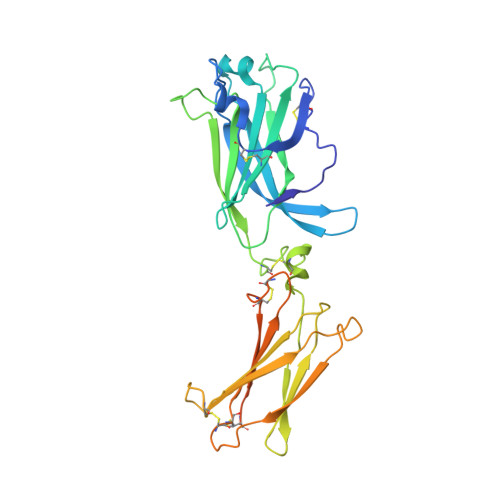
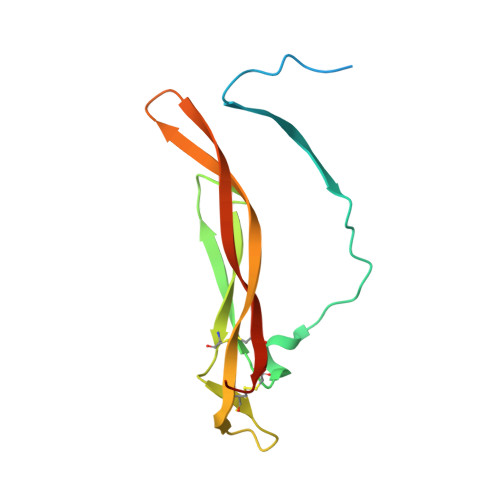
The human IL-17A/F heterodimer: a two-faced cytokine with unique receptor recognition properties.
Goepfert, A., Lehmann, S., Wirth, E., Rondeau, J.M.(2017) Sci Rep 7: 8906-8906
- PubMed: 28827714
- DOI: https://doi.org/10.1038/s41598-017-08360-9
- Primary Citation of Related Structures:
5N92, 5NAN - PubMed Abstract:
IL-17A and IL-17F are prominent members of the IL-17 family of cytokines that regulates both innate and adaptive immunity. IL-17A has been implicated in chronic inflammatory and autoimmune diseases, and anti-IL-17A antibodies have shown remarkable clinical efficacy in psoriasis and psoriatic arthritis patients. IL-17A and IL-17F are homodimeric cytokines that can also form the IL-17A/F heterodimer whose precise role in health and disease remains elusive. All three cytokines signal through the assembly of a ternary complex with the IL-17RA and IL-17RC receptors. Here we report the X-ray analysis of the human IL-17A/F heterodimer that reveals a two-faced cytokine closely mimicking IL-17A as well as IL-17F. We also present the crystal structure of its complex with the IL-17RA receptor. Unexpectedly in view of the much higher affinity of this receptor toward IL-17A, we find that IL-17RA is bound to the "F-face" of the heterodimer in the crystal. Using site-directed mutagenesis, we then demonstrate that IL-17RA can also bind to the "A-face" of IL-17A/F with similar affinity. Further, we show that IL-17RC does not discriminate between the two faces of the cytokine heterodimer either, thus enabling the formation of two topologically-distinct heterotrimeric complexes with potentially different signaling properties.
Organizational Affiliation:
Novartis Institutes for BioMedical Research, Novartis Pharma AG, CH-4002, Basel, Switzerland.