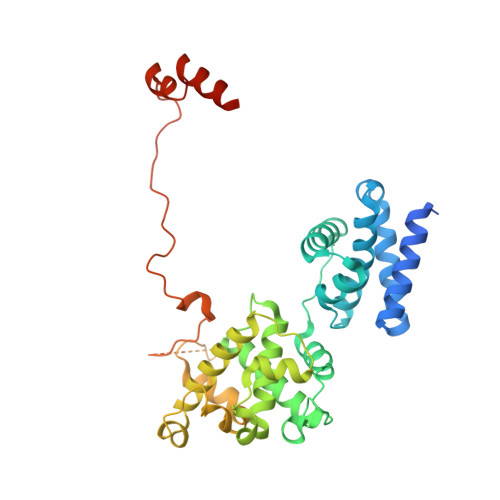
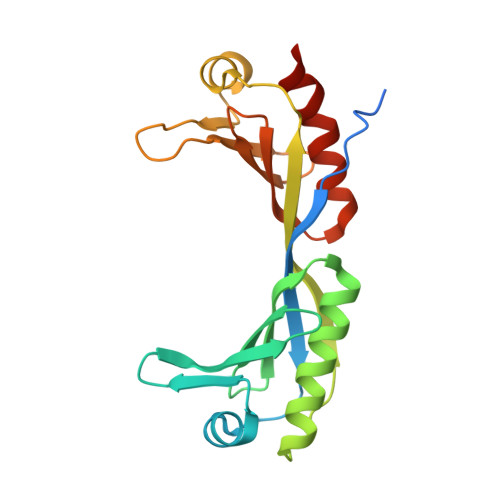
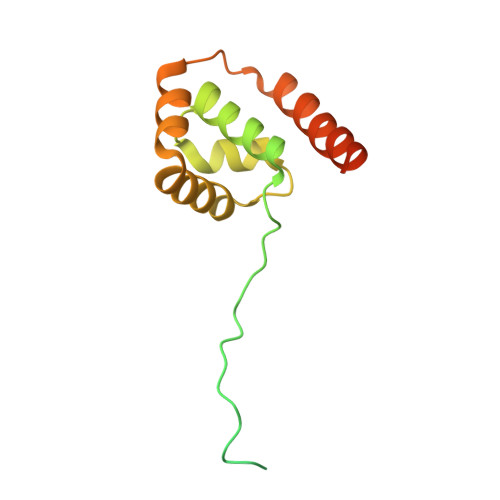
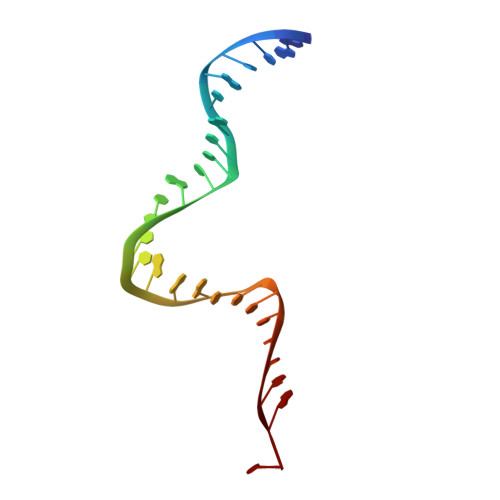Molecular mechanisms of Bdp1 in TFIIIB assembly and RNA polymerase III transcription initiation.
Gouge, J., Guthertz, N., Kramm, K., Dergai, O., Abascal-Palacios, G., Satia, K., Cousin, P., Hernandez, N., Grohmann, D., Vannini, A.(2017) Nat Commun 8: 130-130
- PubMed: 28743884
- DOI: https://doi.org/10.1038/s41467-017-00126-1
- Primary Citation of Related Structures:
5N9G - PubMed Abstract:
Initiation of gene transcription by RNA polymerase (Pol) III requires the activity of TFIIIB, a complex formed by Brf1 (or Brf2), TBP (TATA-binding protein), and Bdp1. TFIIIB is required for recruitment of Pol III and to promote the transition from a closed to an open Pol III pre-initiation complex, a process dependent on the activity of the Bdp1 subunit. Here, we present a crystal structure of a Brf2-TBP-Bdp1 complex bound to DNA at 2.7 Å resolution, integrated with single-molecule FRET analysis and in vitro biochemical assays. Our study provides a structural insight on how Bdp1 is assembled into TFIIIB complexes, reveals structural and functional similarities between Bdp1 and Pol II factors TFIIA and TFIIF, and unravels essential interactions with DNA and with the upstream factor SNAPc. Furthermore, our data support the idea of a concerted mechanism involving TFIIIB and RNA polymerase III subunits for the closed to open pre-initiation complex transition.Transcription initiation by RNA polymerase III requires TFIIIB, a complex formed by Brf1/Brf2, TBP and Bdp1. Here, the authors describe the crystal structure of a Brf2-TBP-Bdp1 complex bound to a DNA promoter and characterize the role of Bdp1 in TFIIIB assembly and pre-initiation complex formation.
Organizational Affiliation:
The Institute of Cancer Research, London, SW7 3RP, UK.